 |
QMCPACK
|
 |
QMCPACK
|
helper functions for EinsplineSetBuilder More...
Namespaces | |
| C2C | |
| C2R | |
| compute | |
| Concurrency | |
| cuBLAS | |
| interface to cuBLAS calls for different data types S/C/D/Z | |
| cuBLAS_LU | |
| cuBLAS_MFs | |
| Implement selected batched BLAS1/2 calls using CUDA for different data types S/C/D/Z. | |
| CUDA | |
| interface to cuBLAS_inhouse calls for different data types S/C/D/Z | |
| cusolver | |
| interface to cusolver calls for different data types S/C/D/Z | |
| estimatorinput | |
| ewaldref | |
| hdf | |
| input | |
| MatrixOperators | |
| modernstrutil | |
| prevent clash with string_utils.h | |
| ompBLAS | |
| Implement selected batched and non-batched BLAS2 calls using OpenMP offload for different data types S/C/D/Z 1) column major like the BLAS fortran API 2) all the functions are synchronous, expected to be changed to asynchronous in the future. | |
| rocsolver | |
| interface to rocsolver calls for different data types S/C/D/Z | |
| simd | |
| spoinfo | |
| SYCL | |
| syclBLAS | |
| syclSolver | |
| testing | |
| SpaceGrid refactored for use with batched estimator design NE should be dropped when QMCHamiltonian/SpaceGrid has been deleted. | |
| Units | |
Typedefs | |
| template<typename T > | |
| using | IsComplex = std::enable_if_t< IsComplex_t< T >::value, bool > |
| template<typename T > | |
| using | IsReal_t = std::is_floating_point< T > |
| template<typename T > | |
| using | IsReal = std::enable_if_t< std::is_floating_point< T >::value, bool > |
| template<typename T > | |
| using | RealAlias = typename RealAlias_impl< T >::value_type |
| If you have a function templated on a value that can be real or complex and you need to get the base Real type if its complex or just the real. More... | |
| template<typename TREAL , typename TREF , typename = std::enable_if_t<std::is_floating_point<TREAL>::value>> | |
| using | ValueAlias = typename ValueAlias_impl< TREAL, TREF >::value_type |
| If you need to make a value type of a given precision based on a reference value type set the desired POD float point type as TREAL and set the reference type as TREF. More... | |
| template<typename T > | |
| using | RngValueType = typename std::disjunction< OnTypesEqual< T, float, float >, OnTypesEqual< T, double, double >, OnTypesEqual< T, std::complex< float >, float >, OnTypesEqual< T, std::complex< double >, double >, default_type< void > >::type |
| Get the right type for rng in case of complex T. More... | |
| template<typename T > | |
| using | OffloadPinnedAllocator = OMPallocator< T, PinnedAlignedAllocator< T > > |
| using | ChronoClock = std::chrono::system_clock |
| using | timer_id_t = unsigned char |
| using | StackKey = StackKeyParam< 2 > |
| using | NewTimer = TimerType< ChronoClock > |
| using | FakeTimer = TimerType< FakeChronoClock > |
| using | ScopedTimer = ScopeGuard< NewTimer > |
| using | ScopedFakeTimer = ScopeGuard< FakeTimer > |
| using | RandomGenerator = StdRandom< OHMMS_PRECISION_FULL > |
| using | ParseCaseVector_t = vector< ParseCase > |
| using | FakeTimerManager = TimerManager< FakeTimer > |
| template<class T > | |
| using | TimerNameList_t = std::vector< TimerIDName_t< T > > |
| using | TimerList_t = TimerList< NewTimer > |
| using | real_type = OHMMS_PRECISION |
| using | RealType = double |
| template<class Allocator > | |
| using | IsHostSafe = std::enable_if_t< qmc_allocator_traits< Allocator >::is_host_accessible > |
| template<class Allocator > | |
| using | IsNotHostSafe = std::enable_if_t<!qmc_allocator_traits< Allocator >::is_host_accessible > |
| template<class Allocator > | |
| using | IsDualSpace = std::enable_if_t< qmc_allocator_traits< Allocator >::is_dual_space > |
| template<class T , size_t ALIGN = QMC_SIMD_ALIGNMENT> | |
| using | aligned_allocator = Mallocator< T, ALIGN > |
| template<class T > | |
| using | aligned_vector = std::vector< T, aligned_allocator< T > > |
| template<typename T > | |
| using | CUDATypeMap = typename std::disjunction< OnTypesEqual< T, float, float >, OnTypesEqual< T, double, double >, OnTypesEqual< T, float *, float * >, OnTypesEqual< T, double *, double * >, OnTypesEqual< T, float **, float ** >, OnTypesEqual< T, double **, double ** >, OnTypesEqual< T, std::complex< double >, cuDoubleComplex >, OnTypesEqual< T, std::complex< float >, cuComplex >, OnTypesEqual< T, std::complex< double > *, cuDoubleComplex * >, OnTypesEqual< T, std::complex< float > **, cuComplex ** >, OnTypesEqual< T, std::complex< double > **, cuDoubleComplex ** >, OnTypesEqual< T, std::complex< float > *, cuComplex * >, OnTypesEqual< T, const std::complex< double > *, const cuDoubleComplex * >, OnTypesEqual< T, const std::complex< float > *, const cuComplex * >, OnTypesEqual< T, const std::complex< float > **, const cuComplex ** >, OnTypesEqual< T, const std::complex< double > **, const cuDoubleComplex ** >, OnTypesEqual< T, const std::complex< float > *const *, const cuComplex *const * >, OnTypesEqual< T, const std::complex< double > *const *, const cuDoubleComplex *const * >, default_type< void > >::type |
| template<typename T > | |
| using | UnpinnedDualAllocator = OffloadAllocator< T > |
| template<typename T > | |
| using | PinnedDualAllocator = OffloadPinnedAllocator< T > |
| template<typename T > | |
| using | DeviceAllocator = OffloadDeviceAllocator< T > |
| template<typename T > | |
| using | OffloadAllocator = OMPallocator< T, aligned_allocator< T > > |
| template<typename T > | |
| using | OffloadDeviceAllocator = aligned_allocator< T > |
| template<typename T > | |
| using | PinnedAllocator = std::allocator< T > |
| The fact that the pinned allocators are not always pinned hurts readability elsewhere. More... | |
| template<typename T , size_t ALIGN = QMC_SIMD_ALIGNMENT> | |
| using | PinnedAlignedAllocator = aligned_allocator< T, ALIGN > |
| using | CPUOMPTargetSelector = PlatformSelector< SelectorKind::CPU_OMPTARGET > |
| using | CPUOMPTargetVendorSelector = PlatformSelector< SelectorKind::CPU_OMPTARGET > |
| template<typename T > | |
| using | hipblasTypeMap = typename std::disjunction< OnTypesEqual< T, float, float >, OnTypesEqual< T, double, double >, OnTypesEqual< T, float *, float * >, OnTypesEqual< T, double *, double * >, OnTypesEqual< T, float **, float ** >, OnTypesEqual< T, double **, double ** >, OnTypesEqual< T, std::complex< double >, hipblasDoubleComplex >, OnTypesEqual< T, std::complex< float >, hipblasComplex >, OnTypesEqual< T, std::complex< double > *, hipblasDoubleComplex * >, OnTypesEqual< T, std::complex< float > **, hipblasComplex ** >, OnTypesEqual< T, std::complex< double > **, hipblasDoubleComplex ** >, OnTypesEqual< T, std::complex< float > *, hipblasComplex * >, OnTypesEqual< T, const std::complex< double > *, const hipblasDoubleComplex * >, OnTypesEqual< T, const std::complex< float > *, const hipblasComplex * >, OnTypesEqual< T, const std::complex< float > **, const hipblasComplex ** >, OnTypesEqual< T, const std::complex< double > **, const hipblasDoubleComplex ** >, OnTypesEqual< T, const std::complex< float > *const *, const hipblasComplex *const * >, OnTypesEqual< T, const std::complex< double > *const *, const hipblasDoubleComplex *const * >, default_type< void > >::type |
| using | vec_t = TinyVector< double, 3 > |
| using | mRealType = EwaldHandler3D::mRealType |
| using | PosType = TinyVector< double, 3 > |
| using | pRealType = qmcplusplus::LRHandlerBase::pRealType |
| using | WP = WalkerProperties::Indexes |
| using | MCPWalker = Walker< QMCTraits, PtclOnLatticeTraits > |
| using | PsiValue = WaveFunctionComponent::PsiValue |
| using | ValueType = LatticeGaussianProduct::ValueType |
| using | GradType = LatticeGaussianProduct::GradType |
| using | ValueVector = OrbitalSetTraits< ValueType >::ValueVector |
| using | BasisSet_t = LCAOrbitalSet::basis_type |
| using | opt_variables_type = optimize::VariableSet |
| using | QuantumNumberType = TinyVector< int, 4 > |
| using | SPOSetPtr = SPOSet * |
| template<typename T > | |
| using | OffloadPinnedMatrix = Matrix< T, PinnedDualAllocator< T > > |
| template<typename T > | |
| using | OffloadPinnedVector = Vector< T, PinnedDualAllocator< T > > |
| using | StdComp = std::complex< double > |
| using | LogValue = std::complex< double > |
| using | ComplexType = QMCTraits::ComplexType |
| using | LogComplexApprox = Catch::Detail::LogComplexApprox |
| using | FakeJasFunctor = FakeFunctor< OHMMS_PRECISION > |
| template<typename DT > | |
| using | OffloadVector = Vector< DT, OffloadPinnedAllocator< DT > > |
| using | DiracDet = DiracDeterminant< DelayedUpdate< QMCTraits::ValueType, QMCTraits::QTFull::ValueType > > |
| using | Return_t = BareKineticEnergy::Return_t |
| using | LRHandlerType = LRCoulombSingleton::LRHandlerType |
| using | GridType = LRCoulombSingleton::GridType |
| using | RadFunctorType = LRCoulombSingleton::RadFunctorType |
| using | Value_t = DensityMatrices1B::Value_t |
| using | Real = ForceBase::Real |
| using | value_type = QMCTraits::FullPrecRealType |
| using | QMCT = QMCTraits |
| using | EstimatorInput = std::variant< std::monostate, MomentumDistributionInput, SpinDensityInput, OneBodyDensityMatricesInput, SelfHealingOverlapInput, MagnetizationDensityInput, PerParticleHamiltonianLoggerInput > |
| using | EstimatorInputs = std::vector< EstimatorInput > |
| using | ScalarEstimatorInput = std::variant< std::monostate, LocalEnergyInput, CSLocalEnergyInput, RMCLocalEnergyInput > |
| using | ScalarEstimatorInputs = std::vector< ScalarEstimatorInput > |
| using | SPOMap = SPOSet::SPOMap |
| using | input = testing::ValidSpinDensityInput |
| using | TraceInt = long |
| using | TraceReal = OHMMS_PRECISION |
| using | TraceComp = std::complex< TraceReal > |
| using | FullPrecReal = DMCRefEnergy::FullPrecReal |
| using | FullPrecValueType = qmcplusplus::QMCTraits::FullPrecValueType |
| using | FullPrecRealType = HamiltonianRef::FullPrecRealType |
| using | IndexType = QMCTraits::IndexType |
Functions | |
| float | real (const float &c) |
| real part of a scalar. Cannot be replaced by std::real due to AFQMC specific needs. More... | |
| double | real (const double &c) |
| float | real (const std::complex< float > &c) |
| double | real (const std::complex< double > &c) |
| float | imag (const float &c) |
| imaginary part of a scalar. Cannot be replaced by std::imag due to AFQMC specific needs. More... | |
| double | imag (const double &c) |
| float | imag (const std::complex< float > &c) |
| double | imag (const std::complex< double > &c) |
| float | conj (const float &c) |
| Workaround to allow conj on scalar to return real instead of complex. More... | |
| double | conj (const double &c) |
| std::complex< float > | conj (const std::complex< float > &c) |
| std::complex< double > | conj (const std::complex< double > &c) |
| void | copy_with_complex_cast (const std::complex< double > &source, std::complex< double > &dest) |
| void | copy_with_complex_cast (const std::complex< double > &source, double &dest) |
| void | copy_with_complex_cast (const std::complex< float > &source, std::complex< float > &dest) |
| void | copy_with_complex_cast (const std::complex< float > &source, float &dest) |
| template<typename T1 , typename T2 , IsReal< T2 > = true> | |
| void | convertToReal (const T1 &in, T2 &out) |
| generic conversion from type T1 to type T2 using implicit conversion More... | |
| template<typename T1 , typename T2 , IsReal< T2 > = true> | |
| void | convertToReal (const std::complex< T1 > &in, T2 &out) |
| specialization of conversion from complex to real More... | |
| template<typename T1 , typename T2 , unsigned D> | |
| void | convertToReal (const TinyVector< T1, D > &in, TinyVector< T2, D > &out) |
| template<typename T1 , typename T2 , unsigned D> | |
| void | convertToReal (const Tensor< T1, D > &in, Tensor< T2, D > &out) |
| specialization for D tensory More... | |
| template<typename T1 , typename T2 > | |
| void | convertToReal (const T1 *restrict in, T2 *restrict out, std::size_t n) |
| generic function to convert arrays More... | |
| template<typename T1 , typename T2 > | |
| void | convertToReal (const Vector< T1 > &in, Vector< T2 > &out) |
| specialization for a vector More... | |
| template<typename T1 , typename T2 > | |
| void | convertToReal (const Matrix< T1 > &in, Matrix< T2 > &out) |
| specialization for a vector More... | |
| TEST_CASE ("complex_helper", "[type_traits]") | |
| TEST_CASE ("QMCTypes", "[type_traits]") | |
| TEST_CASE ("makeRefVector", "[type_traits]") | |
| TEST_CASE ("convertUPtrToRefvector", "[type_traits]") | |
| TEST_CASE ("convertPtrToRefvectorSubset", "[type_traits]") | |
| template<class T , class R > | |
| constexpr bool | has (const R &this_one) |
| void | TestTaskOMP (const int ip, int &counter) |
| Openmp generally works but is not guaranteed with std::atomic. More... | |
| TEST_CASE ("ParallelExecutor<OPENMP> function case", "[concurrency]") | |
| TEST_CASE ("ParallelExecutor<OPENMP> lambda case", "[concurrency]") | |
| TEST_CASE ("ParallelExecutor<OPENMP> nested case", "[concurrency]") | |
| void | TestTask (const int ip, std::atomic< int > &counter) |
| TEST_CASE ("ParallelExecutor<STD> function case", "[concurrency]") | |
| TEST_CASE ("ParallelExecutor<STD> lambda case", "[concurrency]") | |
| TEST_CASE ("ParallelExecutor<STD> nested case", "[concurrency]") | |
| TEST_CASE ("UtilityFunctions OpenMP", "[concurrency]") | |
| TEST_CASE ("ConstantSizeMatrix basics", "[containers]") | |
| TEST_CASE ("ConstantSizeMatrix Ohmms integration", "[containers]") | |
| TEST_CASE ("RecordArray basics", "[containers]") | |
| template<class T , unsigned D> | |
| T | trace (const AntiSymTensor< T, D > &) |
| template<class T , unsigned D> | |
| AntiSymTensor< T, D > | transpose (const AntiSymTensor< T, D > &rhs) |
| template<class T , unsigned D> | |
| const AntiSymTensor< T, D > & | operator+ (const AntiSymTensor< T, D > &op) |
| template<class T1 , class T2 , unsigned D> | |
| Tensor< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | dot (const AntiSymTensor< T1, D > &lhs, const AntiSymTensor< T2, D > &rhs) |
| template<class T1 , class T2 , unsigned D> | |
| Tensor< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | dot (const AntiSymTensor< T1, D > &lhs, const Tensor< T2, D > &rhs) |
| template<class T1 , class T2 , unsigned D> | |
| Tensor< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | dot (const Tensor< T1, D > &lhs, const AntiSymTensor< T2, D > &rhs) |
| template<class T1 , class T2 , unsigned D> | |
| Tensor< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | dot (const AntiSymTensor< T1, D > &lhs, const SymTensor< T2, D > &rhs) |
| template<class T1 , class T2 , unsigned D> | |
| Tensor< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | dot (const SymTensor< T1, D > &lhs, const AntiSymTensor< T2, D > &rhs) |
| template<class T1 , class T2 , unsigned D> | |
| TinyVector< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | dot (const TinyVector< T1, D > &lhs, const AntiSymTensor< T2, D > &rhs) |
| template<class T1 , class T2 , unsigned D> | |
| TinyVector< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | dot (const AntiSymTensor< T1, D > &lhs, const TinyVector< T2, D > &rhs) |
| template<class T , unsigned D> | |
| std::ostream & | operator<< (std::ostream &out, const AntiSymTensor< T, D > &rhs) |
| template<class T , class Alloc > | |
| bool | operator== (const Matrix< T, Alloc > &lhs, const Matrix< T, Alloc > &rhs) |
| template<class T , class Alloc > | |
| bool | operator!= (const Matrix< T, Alloc > &lhs, const Matrix< T, Alloc > &rhs) |
| template<class T , typename Alloc > | |
| std::ostream & | operator<< (std::ostream &out, const Matrix< T, Alloc > &rhs) |
| template<class T , typename Alloc > | |
| std::istream & | operator>> (std::istream &is, Matrix< T, Alloc > &rhs) |
| template<class T , typename Alloc , class Op , class RHS > | |
| void | evaluate (Matrix< T, Alloc > &lhs, const Op &op, const Expression< RHS > &rhs) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< OpUnaryMinus, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t > >::Expression_t | operator- (const Matrix< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< OpUnaryPlus, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t > >::Expression_t | operator+ (const Matrix< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< OpBitwiseNot, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t > >::Expression_t | operator~ (const Matrix< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< OpIdentity, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t > >::Expression_t | PETE_identity (const Matrix< T1, C1 > &l) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< UnaryNode< OpCast< T1 >, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | peteCast (const T1 &, const Matrix< T2, C2 > &l) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator+ (const Matrix< T1, C1 > &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator- (const Matrix< T1, C1 > &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator* (const Matrix< T1, C1 > &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator% (const Matrix< T1, C1 > &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator & (const Matrix< T1, C1 > &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator| (const Matrix< T1, C1 > &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator^ (const Matrix< T1, C1 > &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator+ (const Matrix< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator- (const Matrix< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator* (const Matrix< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator% (const Matrix< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator & (const Matrix< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator| (const Matrix< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator^ (const Matrix< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator+ (const Expression< T1 > &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator- (const Expression< T1 > &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator* (const Expression< T1 > &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator% (const Expression< T1 > &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator & (const Expression< T1 > &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator| (const Expression< T1 > &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator^ (const Expression< T1 > &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator+ (const Matrix< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator- (const Matrix< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator* (const Matrix< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator% (const Matrix< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator & (const Matrix< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator| (const Matrix< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator^ (const Matrix< T1, C1 > &l, const T2 &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator+ (const T1 &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator- (const T1 &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator* (const T1 &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator% (const T1 &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator & (const T1 &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator| (const T1 &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Matrix< T2, C2 > >::Leaf_t > >::Expression_t | operator^ (const T1 &l, const Matrix< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class T3 > | |
| MakeReturn< TrinaryNode< FnWhere, typename CreateLeaf< Matrix< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t, typename CreateLeaf< T3 >::Leaf_t > >::Expression_t | where (const Matrix< T1, C1 > &c, const T2 &t, const T3 &f) |
| template<class T1 > | |
| MakeReturn< UnaryNode< OpUnaryMinus, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | operator- (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< OpUnaryPlus, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | operator+ (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< OpBitwiseNot, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | operator~ (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< OpIdentity, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | PETE_identity (const Expression< T1 > &l) |
| template<class T1 , class T2 > | |
| MakeReturn< UnaryNode< OpCast< T1 >, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | peteCast (const T1 &, const Expression< T2 > &l) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator+ (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator- (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator* (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator% (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator & (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator| (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator^ (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator+ (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator- (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator* (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator% (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator & (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator| (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator^ (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator+ (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator- (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator* (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator% (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator & (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator| (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator^ (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 , class T3 > | |
| MakeReturn< TrinaryNode< FnWhere, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t, typename CreateLeaf< T3 >::Leaf_t > >::Expression_t | where (const Expression< T1 > &c, const T2 &t, const T3 &f) |
| template<class T1 , class C1 , class RHS > | |
| Matrix< T1, C1 > & | assign (Matrix< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Matrix< T1, C1 > & | operator+= (Matrix< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Matrix< T1, C1 > & | operator-= (Matrix< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Matrix< T1, C1 > & | operator*= (Matrix< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Matrix< T1, C1 > & | operator%= (Matrix< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Matrix< T1, C1 > & | operator|= (Matrix< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Matrix< T1, C1 > & | operator &= (Matrix< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Matrix< T1, C1 > & | operator^= (Matrix< T1, C1 > &lhs, const RHS &rhs) |
| template<class T , class C , class Op , class RHS > | |
| void | evaluate (Vector< T, C > &lhs, const Op &op, const Expression< RHS > &rhs) |
| template<class T , class Alloc > | |
| bool | operator== (const Vector< T, Alloc > &lhs, const Vector< T, Alloc > &rhs) |
| template<class T , class Alloc > | |
| bool | operator!= (const Vector< T, Alloc > &lhs, const Vector< T, Alloc > &rhs) |
| template<class T , class Alloc > | |
| std::ostream & | operator<< (std::ostream &out, const Vector< T, Alloc > &rhs) |
| template<class T , class Alloc > | |
| std::istream & | operator>> (std::istream &is, Vector< T, Alloc > &rhs) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< FnArcCos, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | acos (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< FnArcSin, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | asin (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< FnArcTan, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | atan (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< FnCeil, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | ceil (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< FnCos, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | cos (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< FnHypCos, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | cosh (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< FnExp, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | exp (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< FnFabs, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | abs (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< FnFloor, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | floor (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< FnLog, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | log (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< FnLog10, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | log10 (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< FnSin, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | sin (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< FnHypSin, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | sinh (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< FnSqrt, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | sqrt (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< FnTan, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | tan (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< FnHypTan, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | tanh (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< OpUnaryMinus, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | operator- (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< OpUnaryPlus, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | operator+ (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< OpBitwiseNot, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | operator~ (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< OpIdentity, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | PETE_identity (const Vector< T1, C1 > &l) |
| template<class T1 , class C1 > | |
| MakeReturn< UnaryNode< OpNot, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t > >::Expression_t | operator! (const Vector< T1, C1 > &l) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< UnaryNode< OpCast< T1 >, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | peteCast (const T1 &, const Vector< T2, C2 > &l) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator+ (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator- (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator* (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpDivide, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator/ (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator% (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator & (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator| (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator^ (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< FnLdexp, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | ldexp (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< FnPow, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | pow (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< FnFmod, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | fmod (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< FnArcTan2, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | atan2 (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpLT, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator< (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpLE, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator<= (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpGT, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator> (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpGE, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator>= (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpEQ, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator== (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpNE, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator!= (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpAnd, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator && (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpOr, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator|| (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpLeftShift, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator<< (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpRightShift, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator>> (const Vector< T1, C1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator+ (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator- (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator* (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpDivide, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator/ (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator% (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator & (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator| (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator^ (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< FnLdexp, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | ldexp (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< FnPow, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | pow (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< FnFmod, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | fmod (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< FnArcTan2, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | atan2 (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpLT, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator< (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpLE, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator<= (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpGT, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator> (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpGE, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator>= (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpEQ, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator== (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpNE, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator!= (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpAnd, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator && (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpOr, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator|| (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpLeftShift, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator<< (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpRightShift, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator>> (const Vector< T1, C1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator+ (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator- (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator* (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpDivide, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator/ (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator% (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator & (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator| (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator^ (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< FnLdexp, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | ldexp (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< FnPow, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | pow (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< FnFmod, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | fmod (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< FnArcTan2, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | atan2 (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpLT, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator< (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpLE, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator<= (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpGT, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator> (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpGE, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator>= (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpEQ, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator== (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpNE, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator!= (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpAnd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator && (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpOr, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator|| (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpLeftShift, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator<< (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpRightShift, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator>> (const Expression< T1 > &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator+ (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator- (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator* (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpDivide, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator/ (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator% (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator & (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator| (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator^ (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< FnLdexp, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | ldexp (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< FnPow, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | pow (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< FnFmod, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | fmod (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< FnArcTan2, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | atan2 (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpLT, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator< (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpLE, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator<= (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpGT, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator> (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpGE, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator>= (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpEQ, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator== (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpNE, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator!= (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpAnd, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator && (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpOr, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator|| (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpLeftShift, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator<< (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class C1 , class T2 > | |
| MakeReturn< BinaryNode< OpRightShift, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator>> (const Vector< T1, C1 > &l, const T2 &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator+ (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator- (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator* (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpDivide, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator/ (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator% (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator & (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator| (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator^ (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< FnLdexp, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | ldexp (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< FnPow, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | pow (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< FnFmod, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | fmod (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< FnArcTan2, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | atan2 (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpLT, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator< (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpLE, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator<= (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpGT, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator> (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpGE, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator>= (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpEQ, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator== (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpNE, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator!= (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpAnd, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator && (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class T2 , class C2 > | |
| MakeReturn< BinaryNode< OpOr, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Vector< T2, C2 > >::Leaf_t > >::Expression_t | operator|| (const T1 &l, const Vector< T2, C2 > &r) |
| template<class T1 , class C1 , class T2 , class T3 > | |
| MakeReturn< TrinaryNode< FnWhere, typename CreateLeaf< Vector< T1, C1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t, typename CreateLeaf< T3 >::Leaf_t > >::Expression_t | where (const Vector< T1, C1 > &c, const T2 &t, const T3 &f) |
| template<class T1 > | |
| MakeReturn< UnaryNode< FnArcCos, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | acos (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< FnArcSin, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | asin (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< FnArcTan, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | atan (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< FnCeil, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | ceil (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< FnCos, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | cos (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< FnHypCos, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | cosh (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< FnExp, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | exp (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< FnFabs, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | abs (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< FnFloor, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | floor (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< FnLog, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | log (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< FnLog10, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | log10 (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< FnSin, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | sin (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< FnHypSin, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | sinh (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< FnSqrt, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | sqrt (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< FnTan, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | tan (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< FnHypTan, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | tanh (const Expression< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< OpNot, typename CreateLeaf< Expression< T1 > >::Leaf_t > >::Expression_t | operator! (const Expression< T1 > &l) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpDivide, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator/ (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator & (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< FnLdexp, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | ldexp (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< FnPow, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | pow (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< FnFmod, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | fmod (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< FnArcTan2, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | atan2 (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpLT, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator< (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpLE, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator<= (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpGT, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator> (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpGE, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator>= (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpEQ, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator== (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpNE, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator!= (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpAnd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator && (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpOr, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator|| (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpLeftShift, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator<< (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpRightShift, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator>> (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpDivide, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator/ (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator & (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< FnLdexp, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | ldexp (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< FnPow, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | pow (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< FnFmod, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | fmod (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< FnArcTan2, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | atan2 (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpLT, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator< (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpLE, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator<= (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpGT, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator> (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpGE, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator>= (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpEQ, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator== (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpNE, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator!= (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpAnd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator && (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpOr, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator|| (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpLeftShift, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator<< (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpRightShift, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator>> (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpDivide, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator/ (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator & (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< FnLdexp, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | ldexp (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< FnPow, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | pow (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< FnFmod, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | fmod (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< FnArcTan2, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | atan2 (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpLT, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator< (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpLE, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator<= (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpGT, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator> (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpGE, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator>= (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpEQ, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator== (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpNE, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator!= (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpAnd, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator && (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpOr, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator|| (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class C1 , class RHS > | |
| Vector< T1, C1 > & | assign (Vector< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Vector< T1, C1 > & | operator+= (Vector< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Vector< T1, C1 > & | operator-= (Vector< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Vector< T1, C1 > & | operator*= (Vector< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Vector< T1, C1 > & | operator/= (Vector< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Vector< T1, C1 > & | operator%= (Vector< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Vector< T1, C1 > & | operator|= (Vector< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Vector< T1, C1 > & | operator &= (Vector< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Vector< T1, C1 > & | operator^= (Vector< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Vector< T1, C1 > & | operator<<= (Vector< T1, C1 > &lhs, const RHS &rhs) |
| template<class T1 , class C1 , class RHS > | |
| Vector< T1, C1 > & | operator>>= (Vector< T1, C1 > &lhs, const RHS &rhs) |
| template<class T , unsigned D> | |
| T | trace (const SymTensor< T, D > &rhs) |
| template<class T , unsigned D> | |
| SymTensor< T, D > | transpose (const SymTensor< T, D > &rhs) |
| template<class T1 , class T2 , unsigned D> | |
| Tensor< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | dot (const SymTensor< T1, D > &lhs, const SymTensor< T2, D > &rhs) |
| template<class T1 , class T2 , unsigned D> | |
| Tensor< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | dot (const SymTensor< T1, D > &lhs, const Tensor< T2, D > &rhs) |
| template<class T1 , class T2 , unsigned D> | |
| Tensor< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | dot (const Tensor< T1, D > &lhs, const SymTensor< T2, D > &rhs) |
| template<class T1 , class T2 , unsigned D> | |
| TinyVector< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | dot (const TinyVector< T1, D > &lhs, const SymTensor< T2, D > &rhs) |
| template<class T1 , class T2 , unsigned D> | |
| TinyVector< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | dot (const SymTensor< T1, D > &lhs, const TinyVector< T2, D > &rhs) |
| template<class T , unsigned D> | |
| std::ostream & | operator<< (std::ostream &out, const SymTensor< T, D > &rhs) |
| template<class T , unsigned D> | |
| T | trace (const Tensor< T, D > &rhs) |
trace 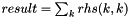 More... More... | |
| template<class T , unsigned D> | |
| Tensor< T, D > | transpose (const Tensor< T, D > &rhs) |
| transpose a tensor More... | |
| template<class T1 , class T2 , unsigned D> | |
| T1 | trace (const Tensor< T1, D > &a, const Tensor< T2, D > &b) |
Tr(a*b), 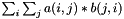 . More... . More... | |
| template<class T , unsigned D> | |
| T | traceAtB (const Tensor< T, D > &a, const Tensor< T, D > &b) |
Tr(a^t *b), 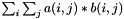 . More... . More... | |
| template<class T1 , class T2 , unsigned D> | |
| BinaryReturn< T1, T2, OpMultiply >::Type_t | traceAtB (const Tensor< T1, D > &a, const Tensor< T2, D > &b) |
Tr(a^t *b), 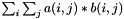 . More... . More... | |
| template<class T1 , class T2 , unsigned D> | |
| Tensor< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | dot (const Tensor< T1, D > &lhs, const Tensor< T2, D > &rhs) |
| Binary Operators. More... | |
| template<class T1 , class T2 , unsigned D> | |
| TinyVector< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | dot (const TinyVector< T1, D > &lhs, const Tensor< T2, D > &rhs) |
Vector-Tensor dot product 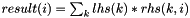 . More... . More... | |
| template<class T1 , class T2 , unsigned D> | |
| TinyVector< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | dot (const Tensor< T1, D > &lhs, const TinyVector< T2, D > &rhs) |
Tensor-Vector dot product 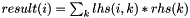 . More... . More... | |
| template<class T , unsigned D> | |
| std::ostream & | operator<< (std::ostream &out, const Tensor< T, D > &rhs) |
Vector-vector outter product 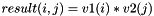 . More... . More... | |
| template<class T , unsigned D> | |
| std::istream & | operator>> (std::istream &is, Tensor< T, D > &rhs) |
| template<class T , unsigned D> | |
| bool | operator== (const Tensor< T, D > &lhs, const Tensor< T, D > &rhs) |
| template<class T , unsigned D> | |
| bool | operator!= (const Tensor< T, D > &lhs, const Tensor< T, D > &rhs) |
| template<class T , unsigned D> | |
| Tensor< T, D >::Type_t | det (const Tensor< T, D > &a) |
| template<class T > | |
| Tensor< T, 1 >::Type_t | det (const Tensor< T, 1 > &a) |
| template<class T > | |
| Tensor< T, 2 >::Type_t | det (const Tensor< T, 2 > &a) |
| template<class T > | |
| Tensor< T, 3 >::Type_t | det (const Tensor< T, 3 > &a) |
| template<class T , unsigned D> | |
| Tensor< T, D > | inverse (const Tensor< T, D > &a) |
| template<class T > | |
| Tensor< T, 1 > | inverse (const Tensor< T, 1 > &a) |
| template<class T > | |
| Tensor< T, 2 > | inverse (const Tensor< T, 2 > &a) |
| template<class T > | |
| Tensor< T, 3 > | inverse (const Tensor< T, 3 > &a) |
| template<class T > | |
| Tensor< T, 3 > | cholesky (const Tensor< T, 3 > &a) |
| TEST_CASE ("array", "[OhmmsPETE]") | |
| TEST_CASE ("array NestedContainers", "[OhmmsPETE]") | |
| TEST_CASE ("Array::data", "[OhmmsPETE]") | |
| TEST_CASE ("Array::dimension sizes constructor", "[OhmmsPETE]") | |
| TEST_CASE ("matrix", "[OhmmsPETE]") | |
| TEST_CASE ("matrix converting assignment", "[OhmmsPETE]") | |
| template<unsigned int D> | |
| void | test_tiny_vector () |
| template<unsigned int D> | |
| void | test_tiny_vector_size_two () |
| TEST_CASE ("tiny vector", "[OhmmsPETE]") | |
| TEST_CASE ("tiny vector operator out", "[OhmmsPETE]") | |
| TEST_CASE ("vector", "[OhmmsPETE]") | |
| TEST_CASE ("Vector simple intializer list", "[OhmmsPETE]") | |
| TEST_CASE ("Vector nested intializer list", "[OhmmsPETE]") | |
| TEST_CASE ("VectorViewer", "[OhmmsPETE]") | |
| TEST_CASE ("NestedContainers", "[OhmmsPETE]") | |
| template<class T1 , unsigned D1, unsigned D2> | |
| TinyMatrix< T1, D2, D1 > | operator! (const TinyMatrix< T1, D1, D2 > &a) |
| template<class T1 > | |
| TinyMatrix< T1, 3, 3 > | operator! (const TinyMatrix< T1, 3, 3 > &a) |
| template<class T1 > | |
| TinyMatrix< T1, 4, 4 > | operator! (const TinyMatrix< T1, 4, 4 > &a) |
| template<class T1 , class T2 , unsigned D> | |
| BinaryReturn< T1, T2, OpMultiply >::Type_t | dot (const TinyVector< T1, D > &lhs, const TinyVector< T2, D > &rhs) |
| template<class T1 , class T2 , unsigned D> | |
| TinyVector< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | cross (const TinyVector< T1, D > &lhs, const TinyVector< T2, D > &rhs) |
| template<class T1 , class T2 , unsigned D> | |
| Tensor< typename BinaryReturn< T1, T2, OpMultiply >::Type_t, D > | outerProduct (const TinyVector< T1, D > &lhs, const TinyVector< T2, D > &rhs) |
| template<class T , unsigned D> | |
| std::ostream & | operator<< (std::ostream &out, const TinyVector< T, D > &rhs) |
| template<class T , unsigned D> | |
| std::istream & | operator>> (std::istream &is, TinyVector< T, D > &rhs) |
| template<typename T > | |
| void | PosAoS2SoA (int nrows, int ncols, const T *restrict iptr, int lda, T *restrict out, int ldb) |
| General conversion function from AoS[nrows][ncols] to SoA[ncols][ldb]. More... | |
| template<typename T > | |
| void | PosSoA2AoS (int nrows, int ncols, const T *restrict iptr, int lda, T *restrict out, int ldb) |
| General conversion function from SoA[ncols][ldb] to AoS[nrows][ncols]. More... | |
| TEST_CASE ("vector", "[OhmmsSoA]") | |
| TEST_CASE ("VectorSoaContainer copy constructor", "[OhmmsSoA]") | |
| TEST_CASE ("VectorSoaContainer move constructor", "[OhmmsSoA]") | |
| TEST_CASE ("VectorSoaContainer assignment", "[OhmmsSoA]") | |
| template<class A , class Op , class Tag > | |
| Combine1< A, Op, Tag >::Type_t | peteCombine (const A &a, const Op &op, const Tag &t) |
| template<class A , class B , class Op , class Tag > | |
| Combine2< A, B, Op, Tag >::Type_t | peteCombine (const A &a, const B &b, const Op &op, const Tag &t) |
| template<class A , class B , class C , class Op , class Tag > | |
| Combine3< A, B, C, Op, Tag >::Type_t | peteCombine (const A &a, const B &b, const C &c, const Op &op, const Tag &t) |
| template<class Expr , class FTag , class CTag > | |
| ForEach< Expr, FTag, CTag >::Type_t | forEach (const Expr &e, const FTag &f, const CTag &c) |
| TEST_CASE ("pack scalar", "[utilities]") | |
| template<class OPA > | |
| void | testDualAllocator () |
| TEST_CASE ("OhmmsMatrix_VectorSoaContainer_View", "[Integration][Allocators]") | |
| template<typename T , typename ENABLE = std::enable_if_t<!std::is_same<bool, T>::value>> | |
| hid_t | get_h5_datatype (const T &) |
| map C types to hdf5 native types bool is explicit removed due to the fact that it is implementation-dependant More... | |
| BOOSTSUB_H5_DATATYPE (char, H5T_NATIVE_CHAR) | |
| BOOSTSUB_H5_DATATYPE (short, H5T_NATIVE_SHORT) | |
| BOOSTSUB_H5_DATATYPE (int, H5T_NATIVE_INT) | |
| BOOSTSUB_H5_DATATYPE (long, H5T_NATIVE_LONG) | |
| BOOSTSUB_H5_DATATYPE (long long, H5T_NATIVE_LLONG) | |
| BOOSTSUB_H5_DATATYPE (unsigned char, H5T_NATIVE_UCHAR) | |
| BOOSTSUB_H5_DATATYPE (unsigned short, H5T_NATIVE_USHORT) | |
| BOOSTSUB_H5_DATATYPE (unsigned int, H5T_NATIVE_UINT) | |
| BOOSTSUB_H5_DATATYPE (unsigned long, H5T_NATIVE_ULONG) | |
| BOOSTSUB_H5_DATATYPE (unsigned long long, H5T_NATIVE_ULLONG) | |
| BOOSTSUB_H5_DATATYPE (float, H5T_NATIVE_FLOAT) | |
| BOOSTSUB_H5_DATATYPE (double, H5T_NATIVE_DOUBLE) | |
| hdf_path | operator/ (const hdf_path &lhs, const hdf_path &rhs) |
| concatenates two paths with a directory separator More... | |
| hdf_path | operator/ (const hdf_path &lhs, const std::string &rhs) |
| hdf_path | operator/ (const hdf_path &lhs, const char *rhs) |
| bool | operator== (const hdf_path &lhs, const hdf_path &rhs) noexcept |
| Checks whether lhs and rhs are equal. More... | |
| template<typename T , typename IT > | |
| bool | getDataShape (hid_t grp, const std::string &aname, std::vector< IT > &sizes_out) |
| free template function to read the (user) dimension and shape of the dataset. More... | |
| template<typename T > | |
| bool | checkShapeConsistency (hid_t grp, const std::string &aname, int rank, hsize_t *dims) |
| free function to check dimension More... | |
| template<typename T > | |
| bool | h5d_read (hid_t grp, const std::string &aname, T *first, hid_t xfer_plist) |
| return true, if successful More... | |
| bool | h5d_check_existence (hid_t grp, const std::string &aname) |
| template<typename T > | |
| bool | h5d_check_type (hid_t grp, const std::string &aname) |
| template<typename T > | |
| bool | h5d_write (hid_t grp, const std::string &aname, hsize_t ndims, const hsize_t *dims, const T *first, hid_t xfer_plist) |
| template<typename T > | |
| bool | h5d_read (hid_t grp, const std::string &aname, hsize_t ndims, const hsize_t *gcounts, const hsize_t *counts, const hsize_t *offsets, T *first, hid_t xfer_plist) |
| return true, if successful More... | |
| template<typename T > | |
| bool | h5d_write (hid_t grp, const std::string &aname, hsize_t ndims, const hsize_t *gcounts, const hsize_t *counts, const hsize_t *offsets, const T *first, hid_t xfer_plist) |
| template<typename T > | |
| bool | h5d_read (hid_t grp, const std::string &aname, hsize_t ndims, const hsize_t *gcounts, const hsize_t *counts, const hsize_t *offsets, hsize_t mem_ndims, const hsize_t *mem_gcounts, const hsize_t *mem_counts, const hsize_t *mem_offsets, T *first, hid_t xfer_plist) |
| return true, if successful More... | |
| template<typename T > | |
| bool | h5d_write (hid_t grp, const std::string &aname, hsize_t ndims, const hsize_t *gcounts, const hsize_t *counts, const hsize_t *offsets, hsize_t mem_ndims, const hsize_t *mem_gcounts, const hsize_t *mem_counts, const hsize_t *mem_offsets, const T *first, hid_t xfer_plist) |
| template<typename T > | |
| bool | h5d_append (hid_t grp, const std::string &aname, hsize_t ¤t, hsize_t ndims, const hsize_t *const dims, const T *const first, hsize_t chunk_size=1, hid_t xfer_plist=H5P_DEFAULT) |
| std::ostream & | operator<< (std::ostream &out, const HDFVersion &v) |
| std::istream & | operator>> (std::istream &is, HDFVersion &v) |
| template bool | approxEquality< float > (float val_a, float val_b, std::optional< double > eps) |
| template bool | approxEquality< std::complex< float > > (std::complex< float > val_a, std::complex< float > val_b, std::optional< double > eps) |
| template bool | approxEquality< double > (double val_a, double val_b, std::optional< double > eps) |
| template bool | approxEquality< std::complex< double > > (std::complex< double > val_a, std::complex< double > val_b, std::optional< double > eps) |
| template<typename T , IsComplex< T > = true> | |
| bool | approxEquality (T val_a, T val_b, std::optional< double > eps) |
| template<class M1 , class M2 > | |
| CheckMatrixResult | checkMatrix (M1 &a_mat, M2 &b_mat, const bool check_all=false, std::optional< const double > eps=std::nullopt) |
| This function checks equality a_mat and b_mat elements M1, M2 need to have their element type declared M1::value_type and have an operator(i,j) accessor. More... | |
| template<class T , unsigned D> | |
| std::ostream & | operator<< (std::ostream &out, const NativePrint< TinyVector< T, D >> &np_vec) |
| template<class T > | |
| std::ostream & | operator<< (std::ostream &out, const NativePrint< std::vector< T >> &np_vec) |
| template<> | |
| std::ostream & | operator<< (std::ostream &out, const NativePrint< std::vector< bool >> &np_vec) |
| template<class T > | |
| std::ostream & | operator<< (std::ostream &out, const NativePrint< Vector< T >> &np_vec) |
| template<class IT > | |
| void | delete_iter (IT first, IT last) |
| delete the pointers in [first,last) More... | |
| template<typename IT1 , typename IT2 > | |
| void | accumulate_elements (IT1 first, IT1 last, IT2 res) |
| template<class T > | |
| ReportEngine & | operator<< (ReportEngine &o, const T &val) |
| template<typename T > | |
| std::ostream & | operator<< (std::ostream &out, const std::vector< T > &rhs) |
| collapsed vector printout [3, 3, 2, 2] is printed as [3(x2), 2(x2)] More... | |
| std::string | strip (const std::string &s) |
| bool | whitespace (char c) |
| std::vector< std::string > | split (const std::string &s) |
| std::vector< std::string > | split (const std::string &s, const std::string &pattern) |
| int | string2int (const std::string &s) |
| double | string2real (const std::string &s) |
| std::string | int2string (const int &i) |
| std::string | real2string (const double &r) |
| bool | string2bool (const std::string &s) |
| template<class T > | |
| std::vector< T > | convertStrToVec (const std::string &s) |
| extract the contents of a string to a vector of something. separator is white spaces. More... | |
| std::istream & | operator>> (std::istream &is, astring &rhs) |
| std::ostream & | operator<< (std::ostream &os, const astring &rhs) |
| bool | operator== (const astring &lhs, const astring &rhs) |
| TEST_CASE ("checkMatrix_OhmmsMatrix_real", "[utilities][for_testing]") | |
| TEST_CASE ("checkMatrix_real", "[utilities][for_testing]") | |
| TEST_CASE ("RandomForTest_fillBufferRng_real", "[utilities][for_testing]") | |
| TEST_CASE ("RandomForTest_fillVectorRngReal", "[utilities][for_testing]") | |
| TEST_CASE ("RandomForTest_getVecRngReal", "[utilities][for_testing]") | |
| TEST_CASE ("RandomForTest_fillBufferRngComplex", "[utilities][for_testing]") | |
| TEST_CASE ("RandomForTest_fillVecRngComplex", "[utilities][for_testing]") | |
| TEST_CASE ("RandomForTest_getVecRngComplex", "[utilities][for_testing]") | |
| TEST_CASE ("RandomForTest_call_operator", "[utilities][for_testing]") | |
| TEST_CASE ("FakeRandom determinism", "[utilities]") | |
| TEST_CASE ("FakeRandom set_value", "[utilities]") | |
| TEST_CASE ("FakeRandom read write", "[utilities]") | |
| TEST_CASE ("FakeRandom noops", "[utilities]") | |
| TEST_CASE ("FakeRandom clone", "[utilities]") | |
| TEST_CASE ("ModernStringUtils_strToLower", "[utilities]") | |
| TEST_CASE ("ModernStringUtils_split", "[utilities]") | |
| TEST_CASE ("ModernStringUtils_string2Real", "[utilities]") | |
| TEST_CASE ("ModernStringUtils_string2Int", "[utilities]") | |
| TEST_CASE ("ModernStringUtils_strip", "[utilities]") | |
| void | reset_string_output () |
| void | init_string_output () |
| TEST_CASE ("OutputManager basic", "[utilities]") | |
| TEST_CASE ("OutputManager output", "[utilities]") | |
| TEST_CASE ("OutputManager pause", "[utilities]") | |
| TEST_CASE ("OutputManager shutoff", "[utilities]") | |
| TEST_CASE ("parsewords_empty", "[utilities]") | |
| TEST_CASE ("parsewords", "[utilities]") | |
| TEST_CASE ("readLine", "[utilities]") | |
| TEST_CASE ("getwords", "[utilities]") | |
| TEST_CASE ("getwordsWithMergedNumbers", "[utilities]") | |
| void | print_vector (vector< int > &out) |
| TEST_CASE ("FairDivideLow_one", "[utilities]") | |
| TEST_CASE ("FairDivideLow_two", "[utilities]") | |
| TEST_CASE ("FairDivideLow_three", "[utilities]") | |
| TEST_CASE ("FairDivideLow_four", "[utilities]") | |
| TEST_CASE ("FairDivideAligned", "[utilities]") | |
| TEST_CASE ("fairDivide_one", "[utilities]") | |
| TEST_CASE ("fairDivide_empties", "[utilities]") | |
| TEST_CASE ("fairDivide_typical", "[utilities]") | |
| TEST_CASE ("fairDivide_even", "[utilities]") | |
| TEST_CASE ("prime number set 32 bit", "[utilities]") | |
| TEST_CASE ("prime number set 64 bit", "[utilities]") | |
| TEST_CASE ("ProjectData", "[ohmmsapp]") | |
| TEST_CASE ("ProjectData::put no series", "[ohmmsapp]") | |
| TEST_CASE ("ProjectData::put with series", "[ohmmsapp]") | |
| TEST_CASE ("ProjectData::TestDriverVersion", "[ohmmsapp]") | |
| TEST_CASE ("ProjectData::TestIsComplex", "[ohmmsapp]") | |
| TEST_CASE ("Resource", "[utilities]") | |
| TEST_CASE ("DummyResource", "[utilities]") | |
| TEST_CASE ("ResourceCollection", "[utilities]") | |
| TEST_CASE ("make_seed", "[utilities]") | |
| TEST_CASE ("RandomNumberControl make_seeds", "[ohmmsapp]") | |
| TEST_CASE ("RandomNumberControl no random in xml", "[ohmmsapp]") | |
| TEST_CASE ("RandomNumberControl random in xml", "[ohmmsapp]") | |
| template<typename T > | |
| FakeChronoClock::duration | convert_to_ns (T in) |
| TEST_CASE ("test_runtime_manager", "[utilities]") | |
| TEST_CASE ("test_loop_timer", "[utilities]") | |
| TEST_CASE ("test_loop_control", "[utilities]") | |
| TEST_CASE ("StdRandom mt19937 determinism", "[utilities]") | |
| TEST_CASE ("StdRandom save and load", "[utilities]") | |
| TEST_CASE ("StdRandom clone", "[utilities]") | |
| TEST_CASE ("StlPrettyPrint", "[utilities]") | |
| TEST_CASE ("string_utils_streamToVec", "[utilities]") | |
| template<class CLOCK > | |
| void | set_total_time (TimerType< CLOCK > *timer, double total_time_input) |
| template<class CLOCK > | |
| void | set_num_calls (TimerType< CLOCK > *timer, long num_calls_input) |
| template<typename T > | |
| double | convert_to_s (T in) |
| TEST_CASE ("test_timer_stack", "[utilities]") | |
| TEST_CASE ("test_timer_scoped", "[utilities]") | |
| TEST_CASE ("test_timer_flat_profile", "[utilities]") | |
| TEST_CASE ("test_timer_flat_profile_same_name", "[utilities]") | |
| TEST_CASE ("test_timer_nested_profile", "[utilities]") | |
| TEST_CASE ("test_timer_nested_profile_two_children", "[utilities]") | |
| TEST_CASE ("test_timer_nested_profile_alt_routes", "[utilities]") | |
| TEST_CASE ("test_timer_nested_profile_collate", "[utilities]") | |
| TEST_CASE ("test setup timers", "[utilities]") | |
| TimerManager< NewTimer > & | getGlobalTimerManager () |
| NewTimer & | createGlobalTimer (const std::string &myname, timer_levels mylevel) |
| int | get_level (const std::string &stack_name) |
| std::string | get_leaf_name (const std::string &stack_name) |
| void | pad_string (const std::string &in, std::string &out, int field_len) |
| template<typename T > | |
| void | bessel_steed_array_cpu (const int lmax, const T x, T *jl) |
| Compute spherical bessel function from 0 to lmax. More... | |
| template<typename T > | |
| void | det_row_update (T *restrict pinv, const T *restrict tv, int m, int rowchanged, T c_ratio, T *restrict temp, T *restrict rcopy) |
| template<typename T > | |
| void | det_col_update (T *restrict pinv, const T *restrict tv, int m, int colchanged, T c_ratio, T *restrict temp, T *restrict rcopy) |
| template<typename T > | |
| void | getRatiosByRowSubstitution (const T *restrict tm_new, const T *restrict r_replaced, T *restrict ratios, int m, int howmany) |
| template<typename T , typename INDARRAY > | |
| void | getRatiosByRowSubstitution (const T *restrict tm_new, const T *restrict r_replaced, T *restrict ratios, int m, const INDARRAY &ind) |
| template<typename T > | |
| void | getRatiosByRowSubstitution_dummy (const T *restrict tm_new, const T *restrict r_replaced, T *restrict ratios, int m, int howmany) |
| template<typename T > | |
| T | getRatioByColSubstitution (const T *restrict pinv, const T *restrict tc, int m, int colchanged) |
| evaluate the determinant ratio with a column substitution More... | |
| template<typename MAT , typename VV > | |
| MAT::value_type | getRatioByColSubstitution (const MAT &pinv, const VV &tc, int colchanged) |
| template<typename MAT , typename VV , typename IV > | |
| MAT::value_type | getRatioByColSubstitution (const MAT &refinv, const VV &tcm, VV &ratios, int m, int colchanged, int r_replaced, IV &ind) |
| evaluate the ratio with a column substitution and multiple row substitutions More... | |
| void | LUFactorization (int n, int m, double *restrict a, int n0, int *restrict piv) |
| LU factorization of double. More... | |
| void | LUFactorization (int n, int m, float *restrict a, const int &n0, int *restrict piv) |
| LU factorization of float. More... | |
| void | LUFactorization (int n, int m, std::complex< double > *restrict a, int n0, int *restrict piv) |
| LU factorization of std::complex<double> More... | |
| void | LUFactorization (int n, int m, std::complex< float > *restrict a, int n0, int *restrict piv) |
| LU factorization of complex<float> More... | |
| void | InvertLU (int n, double *restrict a, int n0, int *restrict piv, double *restrict work, int n1) |
| Inversion of a double matrix after LU factorization. More... | |
| void | InvertLU (const int &n, float *restrict a, const int &n0, int *restrict piv, float *restrict work, const int &n1) |
| Inversion of a float matrix after LU factorization. More... | |
| void | InvertLU (int n, std::complex< double > *restrict a, int n0, int *restrict piv, std::complex< double > *restrict work, int n1) |
| Inversion of a std::complex<double> matrix after LU factorization. More... | |
| void | InvertLU (int n, std::complex< float > *restrict a, int n0, int *restrict piv, std::complex< float > *restrict work, int n1) |
| Inversion of a complex<float> matrix after LU factorization. More... | |
| template<class T > | |
| T | Invert (T *restrict x, int n, int m, T *restrict work, int *restrict pivot) |
| inverse a matrix More... | |
| template<class T > | |
| T | Determinant (T *restrict x, int n, int m, int *restrict pivot) |
| determinant of a matrix More... | |
| template<class T > | |
| T | Invert (T *restrict x, int n, int m) |
| inverse a matrix More... | |
| template<class T , class T1 > | |
| void | InvertWithLog (T *restrict x, int n, int m, T *restrict work, int *restrict pivot, std::complex< T1 > &logdet) |
| template<class MatrixA > | |
| MatrixA::value_type | invert_matrix (MatrixA &M, bool getdet=true) |
| invert a matrix More... | |
| template<typename MatA , typename VecB > | |
| MatA::value_type | DetRatioByRow (const MatA &Minv, const VecB &newv, int rowchanged) |
| determinant ratio with a row substitution More... | |
| template<typename MatA , typename VecB > | |
| MatA::value_type | DetRatioByColumn (const MatA &Minv, const VecB &newv, int colchanged) |
| determinant ratio with a column substitution More... | |
| template<typename T , typename ALLOC > | |
| void | InverseUpdateByRow (Matrix< T, ALLOC > &Minv, Vector< T, ALLOC > &newrow, Vector< T, ALLOC > &rvec, Vector< T, ALLOC > &rvecinv, int rowchanged, T c_ratio) |
| update a inverse matrix by a row substitution More... | |
| template<typename T , typename ALLOC > | |
| void | InverseUpdateByColumn (Matrix< T, ALLOC > &Minv, Vector< T, ALLOC > &newcol, Vector< T, ALLOC > &rvec, Vector< T, ALLOC > &rvecinv, int colchanged, T c_ratio) |
| template<typename T > | |
| void | LinearFit (std::vector< T > &y, Matrix< T > &A, std::vector< T > &coefs) |
| template<class T > | |
| std::ostream & | operator<< (std::ostream &out, const OneDimGridBase< T > &rhs) |
| template<typename T > | |
| Tensor< T, 3 > | generateRotationMatrix (T rng1, T rng2, T rng3) |
| Create a random 3D rotation matrix from three random numbers. More... | |
| template<typename T > | |
| T | smoothing (smoothing_functions func_id, T x, T &dx, T &d2x) |
| template float | smoothing (smoothing_functions func_id, float x, float &dx, float &d2x) |
| template double | smoothing (smoothing_functions func_id, double x, double &dx, double &d2x) |
| template<typename T , typename TRESIDUAL > | |
| void | getSplineBound (const T x, const int nmax, int &ind, TRESIDUAL &dx) |
| break x into the integer part and residual part and apply bounds More... | |
| TEST_CASE ("Bessel", "[numerics]") | |
| TEST_CASE ("Cartesian Tensor", "[numerics]") | |
| TEST_CASE ("Cartesian Tensor evaluateAll subset", "[numerics]") | |
| TEST_CASE ("Cartesian Tensor evaluateWithHessian subset", "[numerics]") | |
| TEST_CASE ("Cartesian Tensor evaluateWithThirdDeriv subset", "[numerics]") | |
| TEST_CASE ("Cartesian Tensor evaluateThirdDerivOnly subset", "[numerics]") | |
| TEST_CASE ("Basic Gaussian", "[numerics]") | |
| TEST_CASE ("Gaussian Combo", "[numerics]") | |
| TEST_CASE ("Gaussian Combo P", "[numerics]") | |
| TEST_CASE ("Gaussian Combo D", "[numerics]") | |
| TEST_CASE ("double_1d_grid_functor", "[numerics]") | |
| TEST_CASE ("double_1d_grid_functor_vs_n", "[numerics]") | |
| TEST_CASE ("bracket minimum", "[numerics]") | |
| TEST_CASE ("find minimum", "[numerics]") | |
| TEST_CASE ("spline_function_1", "[numerics]") | |
| TEST_CASE ("spline_function_2", "[numerics]") | |
| TEST_CASE ("spline_function_3", "[numerics]") | |
| TEST_CASE ("spline_function_4", "[numerics]") | |
| TEST_CASE ("spline_function_5", "[numerics]") | |
| TEST_CASE ("one_dim_cubic_spline_1", "[numerics]") | |
| TEST_CASE ("test oneDimCubicSplineLinearGrid", "[numerics]") | |
| TEST_CASE ("CheckSphericalIntegration", "[numerics]") | |
| TEMPLATE_TEST_CASE ("RandomRotationMatrix", "[numerics]", float, double) | |
| TEST_CASE ("SoA Cartesian Tensor", "[numerics]") | |
| TEST_CASE ("SoA Cartesian Tensor evaluateVGL subset", "[numerics]") | |
| TEST_CASE ("SoA Cartesian Tensor evaluateVGH subset", "[numerics]") | |
| template<typename T > | |
| void | test_spline_bounds () |
| TEST_CASE ("getSplineBound double", "[numerics]") | |
| TEST_CASE ("getSplineBound float", "[numerics]") | |
| TEST_CASE ("stdlib round", "[numerics]") | |
| TEST_CASE ("transform2gridfunctor", "[numerics]") | |
| TEST_CASE ("Legendre", "[numerics]") | |
| TEST_CASE ("Spherical Harmonics", "[numerics]") | |
| TEST_CASE ("Spherical Harmonics Many", "[numerics]") | |
| TEST_CASE ("Derivatives of Spherical Harmonics", "[numerics]") | |
| TEST_CASE ("Spherical Harmonics Wrapper", "[numerics]") | |
| TEST_CASE ("Cartesian derivatives of Spherical Harmonics", "[numerics]") | |
| template<typename T > | |
| T | LegendrePll (int l, T x) |
| template<typename T > | |
| T | LegendrePlm (int l, int m, T x) |
| template<typename T > | |
| std::complex< T > | Ylm (int l, int m, const TinyVector< T, 3 > &r) |
| calculates Ylm param[in] l angular momentum param[in] m magnetic quantum number param[in] r position vector. More... | |
| template<typename T > | |
| void | derivYlmSpherical (const int l, const int m, const TinyVector< T, 3 > &r, std::complex< T > &theta_deriv, std::complex< T > &phi_deriv, const bool conj) |
| calculate the derivative of a Ylm with respect to theta and phi param[in] l: angular momentum param[in] m: magnetic quantum number param[in] r: cartesian position align with [z,x,y]. More... | |
| template<typename T > | |
| std::complex< T > | sphericalHarmonic (const int l, const int m, const TinyVector< T, 3 > &r) |
| wrapper for Ylm, which can take any normal position vector as an argument param[in] l angular momentum param[in] m magnetic quantum number param[in] r is a position vector. More... | |
| template<typename T > | |
| void | sphericalHarmonicGrad (const int l, const int m, const TinyVector< T, 3 > &r, TinyVector< std::complex< T >, 3 > &grad) |
| get cartesian derivatives of spherical Harmonics. More... | |
| bool | isnan (float) |
| return true if the value is NaN. More... | |
| bool | isnan (double a) |
| bool | isfinite (float) |
| return true if the value is finite. More... | |
| bool | isfinite (double a) |
| bool | isinf (float) |
| return true if the value is Inf. More... | |
| bool | isinf (double a) |
| template<typename T > | |
| void | sincos (T a, T *restrict s, T *restrict c) |
| sincos function wrapper More... | |
| int | pow (int i, int n) |
| return i^n More... | |
| template<typename T , typename = typename std::enable_if<std::is_floating_point<T>::value>::type> | |
| bool | iszero (T a) |
| template<class T1 , size_t ALIGN1, class T2 , size_t ALIGN2> | |
| bool | operator== (const Mallocator< T1, ALIGN1 > &, const Mallocator< T2, ALIGN2 > &) |
| template<class T1 , size_t ALIGN1, class T2 , size_t ALIGN2> | |
| bool | operator!= (const Mallocator< T1, ALIGN1 > &, const Mallocator< T2, ALIGN2 > &) |
| std::atomic< size_t > | CUDAallocator_device_mem_allocated (0) |
| size_t | getCUDAdeviceMemAllocated () |
| template<class T1 , class T2 > | |
| bool | operator== (const CUDAManagedAllocator< T1 > &, const CUDAManagedAllocator< T2 > &) |
| template<class T1 , class T2 > | |
| bool | operator!= (const CUDAManagedAllocator< T1 > &, const CUDAManagedAllocator< T2 > &) |
| template<class T1 , class T2 > | |
| bool | operator== (const CUDAAllocator< T1 > &, const CUDAAllocator< T2 > &) |
| template<class T1 , class T2 > | |
| bool | operator!= (const CUDAAllocator< T1 > &, const CUDAAllocator< T2 > &) |
| template<class T1 , class T2 > | |
| bool | operator== (const CUDAHostAllocator< T1 > &, const CUDAHostAllocator< T2 > &) |
| template<class T1 , class T2 > | |
| bool | operator!= (const CUDAHostAllocator< T1 > &, const CUDAHostAllocator< T2 > &) |
| template<typename T > | |
| void | CUDAfill_n (T *ptr, size_t n, const T &value) |
| fill device memory with a given value. More... | |
| template void | CUDAfill_n< int > (int *ptr, size_t n, const int &value) |
| template void | CUDAfill_n< size_t > (size_t *ptr, size_t n, const size_t &value) |
| template void | CUDAfill_n< float > (float *ptr, size_t n, const float &value) |
| template void | CUDAfill_n< double > (double *ptr, size_t n, const double &value) |
| template void | CUDAfill_n< std::complex< float > > (std::complex< float > *ptr, size_t n, const std::complex< float > &value) |
| template void | CUDAfill_n< std::complex< double > > (std::complex< double > *ptr, size_t n, const std::complex< double > &value) |
| template<typename T > | |
| CUDATypeMap< T > | castCUDAType (T var) |
| std::atomic< size_t > | dual_device_mem_allocated (0) |
| size_t | getDualDeviceMemAllocated () |
| int | determineDefaultDeviceNum (int num_devices, int rank_id, int num_ranks) |
| distribute MPI ranks among devices More... | |
| std::ostream & | app_summary () |
| std::ostream & | app_log () |
| std::ostream & | app_error () |
| std::ostream & | app_warning () |
| std::ostream & | app_debug_stream () |
| void | print_mem (const std::string &title, std::ostream &log) |
| std::atomic< size_t > | OMPallocator_device_mem_allocated (0) |
| size_t | getOMPdeviceMemAllocated () |
| template<typename T > | |
| T * | getOffloadDevicePtr (T *host_ptr) |
| template<typename T > | |
| hipblasTypeMap< T > | casthipblasType (T var) |
| std::atomic< size_t > | SYCLallocator_device_mem_allocated (0) |
| size_t | getSYCLdeviceMemAllocated () |
| template<class T1 , class T2 > | |
| bool | operator== (const SYCLSharedAllocator< T1 > &, const SYCLSharedAllocator< T2 > &) |
| template<class T1 , class T2 > | |
| bool | operator!= (const SYCLSharedAllocator< T1 > &, const SYCLSharedAllocator< T2 > &) |
| template<class T1 , class T2 > | |
| bool | operator== (const SYCLAllocator< T1 > &, const SYCLAllocator< T2 > &) |
| template<class T1 , class T2 > | |
| bool | operator!= (const SYCLAllocator< T1 > &, const SYCLAllocator< T2 > &) |
| template<class T1 , class T2 > | |
| bool | operator== (const SYCLHostAllocator< T1 > &, const SYCLHostAllocator< T2 > &) |
| template<class T1 , class T2 > | |
| bool | operator!= (const SYCLHostAllocator< T1 > &, const SYCLHostAllocator< T2 > &) |
| sycl::queue & | getSYCLDefaultDeviceDefaultQueue () |
| return a reference to the per-device default queue More... | |
| sycl::queue | createSYCLInOrderQueueOnDefaultDevice () |
| create an in-order queue using the default device More... | |
| sycl::queue | createSYCLQueueOnDefaultDevice () |
| create a out-of-order queue using the default device More... | |
| size_t | getSYCLdeviceFreeMem () |
| query free memory on the default device More... | |
| TEST_CASE ("Aligned allocator", "[numerics]") | |
| template<unsigned int N, typename T > | |
| void | test_e2iphi () |
| TEST_CASE ("e2iphi", "[numerics]") | |
| template<typename T > | |
| void | test_isnan () |
| TEST_CASE ("isnan", "[numerics]") | |
| TEST_CASE ("CUDA_allocators", "[CUDA]") | |
| TEST_CASE ("OMPclass member", "[OMP]") | |
| TEST_CASE ("OMPdeepcopy", "[OMP]") | |
| TEST_CASE ("OMPmath", "[OMP]") | |
| template<typename T > | |
| void | test_gemm (const int M, const int N, const int K, const char transa, const char transb) |
| TEST_CASE ("ompBLAS gemm", "[OMP]") | |
| template<typename T > | |
| void | test_gemv (const int M_b, const int N_b, const char trans) |
| template<typename T > | |
| void | test_gemv_batched (const int M_b, const int N_b, const char trans, const int batch_count) |
| TEST_CASE ("ompBLAS gemv", "[OMP]") | |
| TEST_CASE ("ompBLAS gemv notrans", "[OMP]") | |
| template<typename T > | |
| void | test_ger (const int M, const int N) |
| template<typename T > | |
| void | test_ger_batched (const int M, const int N, const int batch_count) |
| TEST_CASE ("ompBLAS ger", "[OMP]") | |
| TEST_CASE ("OMP runtime memory", "[OMP]") | |
| TEST_CASE ("SYCL_allocator", "[SYCL]") | |
| TEST_CASE ("SYCL_host_allocator", "[SYCL]") | |
| TEST_CASE ("OmpBLAS gemv", "[SYCL]") | |
| TEST_CASE ("OmpBLAS gemv notrans", "[SYCL]") | |
| TEST_CASE ("OmpBLAS ger", "[SYCL]") | |
| template<PlatformKind PL, typename T > | |
| void | test_one_gemm (const int M, const int N, const int K, const char transa, const char transb) |
| template<PlatformKind PL> | |
| void | test_gemm_cases () |
| template<PlatformKind PL, typename T > | |
| void | test_one_gemv (const int M_b, const int N_b, const char trans) |
| template<PlatformKind PL> | |
| void | test_gemv_cases () |
| template<PlatformKind PL, typename T > | |
| void | test_one_ger (const int M, const int N) |
| template<PlatformKind PL> | |
| void | test_ger_cases () |
| TEST_CASE ("AccelBLAS", "[BLAS]") | |
| TEST_CASE ("PlatformSelector", "[platform]") | |
| template<typename CT > | |
| void | cancel (CT &r) |
| template<typename T > | |
| void | bcast (T &a, Communicate *comm) |
| TEST_CASE ("test_communicate_split_one", "[message]") | |
| TEST_CASE ("test_communicate_split_two", "[message]") | |
| TEST_CASE ("test_communicate_split_four", "[message]") | |
| void | mpiTestFunctionWrapped (Communicate *comm, std::vector< double > &fake_args) |
| Openmp generally works but is not guaranteed with std::atomic. More... | |
| TEST_CASE ("MPIExceptionWrapper function case", "[Utilities]") | |
| TEST_CASE ("MPIExceptionWrapper lambda case", "[Utilities]") | |
| std::unique_ptr< DistanceTable > | createDistanceTableAA (ParticleSet &s, std::ostream &description) |
| Class to manage multiple DistanceTable objects. More... | |
| std::unique_ptr< DistanceTable > | createDistanceTableAAOMPTarget (ParticleSet &s, std::ostream &description) |
| Adding SymmetricDTD to the list, e.g., el-el distance table. More... | |
| std::unique_ptr< DistanceTable > | createDistanceTable (ParticleSet &s, std::ostream &description) |
| std::unique_ptr< DistanceTable > | createDistanceTableAB (const ParticleSet &s, ParticleSet &t, std::ostream &description) |
| free function create a distable table of s-t More... | |
| std::unique_ptr< DistanceTable > | createDistanceTableABOMPTarget (const ParticleSet &s, ParticleSet &t, std::ostream &description) |
| Adding AsymmetricDTD to the list, e.g., el-el distance table. More... | |
| std::unique_ptr< DistanceTable > | createDistanceTable (const ParticleSet &s, ParticleSet &t, std::ostream &description) |
| constexpr bool | operator & (DTModes x, DTModes y) |
| constexpr DTModes | operator| (DTModes x, DTModes y) |
| constexpr DTModes | operator~ (DTModes x) |
| DTModes & | operator|= (DTModes &x, DTModes y) |
| std::unique_ptr< DynamicCoordinates > | createDynamicCoordinates (const DynamicCoordinateKind kind) |
| create DynamicCoordinates based on kind More... | |
| template<typename T , unsigned D> | |
| void | put2box (ParticleAttrib< TinyVector< T, D >> &inout) |
| inout[i]=inout[i]-floor(inout[i]) More... | |
| template<typename T , unsigned D> | |
| void | put2box (const ParticleAttrib< TinyVector< T, D >> &in, ParticleAttrib< TinyVector< T, D >> &out) |
| out[i]=in[i]-floor(in[i]) More... | |
| template<typename T > | |
| TinyVector< T, 3 > | lower_bound (const TinyVector< T, 3 > &a, const TinyVector< T, 3 > &b) |
| helper function to determine the lower bound of a domain (need to move up) More... | |
| template<typename T > | |
| TinyVector< T, 3 > | upper_bound (const TinyVector< T, 3 > &a, const TinyVector< T, 3 > &b) |
| helper function to determine the upper bound of a domain (need to move up) More... | |
| template<class T , unsigned D> | |
| bool | operator== (const CrystalLattice< T, D > &lhs, const CrystalLattice< T, D > &rhs) |
| template<class T , unsigned D> | |
| bool | operator!= (const CrystalLattice< T, D > &lhs, const CrystalLattice< T, D > &rhs) |
| template<typename T > | |
| bool | found_shorter_base (TinyVector< TinyVector< T, 3 >, 3 > &rb) |
| template<typename T > | |
| void | find_reduced_basis (TinyVector< TinyVector< T, 3 >, 3 > &rb) |
| TEST_CASE ("Crystal_lattice_periodic_bulk", "[lattice]") | |
| Lattice is defined but Open BC is also used. More... | |
| TEST_CASE ("LRBreakupParameters", "[lattice]") | |
| Lattice is defined but Open BC is also used. More... | |
| TEST_CASE ("open_bconds", "[lattice]") | |
| TEST_CASE ("periodic_bulk_bconds", "[lattice]") | |
| Lattice is defined but Open BC is also used. More... | |
| TEST_CASE ("uniform 3D Lattice layout", "[lattice]") | |
| template<typename T > | |
| std::unique_ptr< OneDimCubicSpline< T > > | createSpline4RbyVs_temp (const LRHandlerBase *aLR, T rcut, const LinearGrid< T > &agrid) |
| template<typename T > | |
| std::unique_ptr< OneDimCubicSpline< T > > | createSpline4RbyVsDeriv_temp (const LRHandlerBase *aLR, T rcut, const LinearGrid< T > &agrid) |
| TEST_CASE ("ewald3d", "[lrhandler]") | |
| evalaute bare Coulomb using EwaldHandler3D More... | |
| TEST_CASE ("ewald3d df", "[lrhandler]") | |
| evalaute bare Coulomb derivative using EwaldHandler3D More... | |
| TEST_CASE ("kcontainer at gamma in 3D", "[longrange]") | |
| TEST_CASE ("kcontainer at twist in 3D", "[longrange]") | |
| TEST_CASE ("kcontainer at gamma in 2D", "[longrange]") | |
| TEST_CASE ("kcontainer at twist in 2D", "[longrange]") | |
| TEST_CASE ("dummy", "[lrhandler]") | |
| evalaute bare Coulomb using DummyLRHandler More... | |
| TEST_CASE ("srcoul", "[lrhandler]") | |
| evalaute bare Coulomb in 3D More... | |
| TEST_CASE ("srcoul df", "[lrhandler]") | |
| evalaute bare Coulomb derivative in 3D More... | |
| TEST_CASE ("StructFact", "[lrhandler]") | |
| evalaute bare Coulomb in 3D using LRHandlerTemp More... | |
| TEST_CASE ("temp3d", "[lrhandler]") | |
| evalaute bare Coulomb in 3D using LRHandlerTemp More... | |
| template<class T1 > | |
| MakeReturn< UnaryNode< OpUnaryMinus, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t > >::Expression_t | operator- (const ParticleAttrib< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< OpUnaryPlus, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t > >::Expression_t | operator+ (const ParticleAttrib< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< OpBitwiseNot, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t > >::Expression_t | operator~ (const ParticleAttrib< T1 > &l) |
| template<class T1 > | |
| MakeReturn< UnaryNode< OpIdentity, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t > >::Expression_t | PETE_identity (const ParticleAttrib< T1 > &l) |
| template<class T1 , class T2 > | |
| MakeReturn< UnaryNode< OpCast< T1 >, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | peteCast (const T1 &, const ParticleAttrib< T2 > &l) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator+ (const ParticleAttrib< T1 > &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator- (const ParticleAttrib< T1 > &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator* (const ParticleAttrib< T1 > &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator% (const ParticleAttrib< T1 > &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator & (const ParticleAttrib< T1 > &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator| (const ParticleAttrib< T1 > &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator^ (const ParticleAttrib< T1 > &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator+ (const ParticleAttrib< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator- (const ParticleAttrib< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator* (const ParticleAttrib< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator% (const ParticleAttrib< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator & (const ParticleAttrib< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator| (const ParticleAttrib< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator^ (const ParticleAttrib< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator+ (const Expression< T1 > &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator- (const Expression< T1 > &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator* (const Expression< T1 > &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator% (const Expression< T1 > &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator & (const Expression< T1 > &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator| (const Expression< T1 > &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator^ (const Expression< T1 > &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator+ (const ParticleAttrib< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator- (const ParticleAttrib< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator* (const ParticleAttrib< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator% (const ParticleAttrib< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator & (const ParticleAttrib< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator| (const ParticleAttrib< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator^ (const ParticleAttrib< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpAdd, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator+ (const T1 &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpSubtract, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator- (const T1 &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpMultiply, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator* (const T1 &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpMod, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator% (const T1 &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator & (const T1 &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseOr, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator| (const T1 &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseXor, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< ParticleAttrib< T2 > >::Leaf_t > >::Expression_t | operator^ (const T1 &l, const ParticleAttrib< T2 > &r) |
| template<class T1 , class T2 , class T3 > | |
| MakeReturn< TrinaryNode< FnWhere, typename CreateLeaf< ParticleAttrib< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t, typename CreateLeaf< T3 >::Leaf_t > >::Expression_t | where (const ParticleAttrib< T1 > &c, const T2 &t, const T3 &f) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator & (const Expression< T1 > &l, const Expression< T2 > &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< Expression< T1 > >::Leaf_t, typename CreateLeaf< T2 >::Leaf_t > >::Expression_t | operator & (const Expression< T1 > &l, const T2 &r) |
| template<class T1 , class T2 > | |
| MakeReturn< BinaryNode< OpBitwiseAnd, typename CreateLeaf< T1 >::Leaf_t, typename CreateLeaf< Expression< T2 > >::Leaf_t > >::Expression_t | operator & (const T1 &l, const Expression< T2 > &r) |
| template<class T1 , class RHS > | |
| ParticleAttrib< T1 > & | assign (ParticleAttrib< T1 > &lhs, const RHS &rhs) |
| template<class T1 , class RHS > | |
| ParticleAttrib< T1 > & | operator+= (ParticleAttrib< T1 > &lhs, const RHS &rhs) |
| template<class T1 , class RHS > | |
| ParticleAttrib< T1 > & | operator-= (ParticleAttrib< T1 > &lhs, const RHS &rhs) |
| template<class T1 , class RHS > | |
| ParticleAttrib< T1 > & | operator*= (ParticleAttrib< T1 > &lhs, const RHS &rhs) |
| template<class T1 , class RHS > | |
| ParticleAttrib< T1 > & | operator%= (ParticleAttrib< T1 > &lhs, const RHS &rhs) |
| template<class T1 , class RHS > | |
| ParticleAttrib< T1 > & | operator|= (ParticleAttrib< T1 > &lhs, const RHS &rhs) |
| template<class T1 , class RHS > | |
| ParticleAttrib< T1 > & | operator &= (ParticleAttrib< T1 > &lhs, const RHS &rhs) |
| template<class T1 , class RHS > | |
| ParticleAttrib< T1 > & | operator^= (ParticleAttrib< T1 > &lhs, const RHS &rhs) |
| template<class T , class Op , class RHS > | |
| void | evaluate (ParticleAttrib< T > &lhs, const Op &op, const Expression< RHS > &rhs) |
| template<class T > | |
| std::ostream & | operator<< (std::ostream &out, const ParticleAttrib< T > &rhs) |
| template<class T , unsigned D> | |
| T * | get_first_address (ParticleAttrib< TinyVector< T, D >> &a) |
| template<class T , unsigned D> | |
| T * | get_last_address (ParticleAttrib< TinyVector< T, D >> &a) |
| template<class T , unsigned D> | |
| void | normalize (ParticleAttrib< TinyVector< T, D >> &pa) |
| template<typename T , unsigned D> | |
| T | Dot (const ParticleAttrib< TinyVector< T, D >> &pa, const ParticleAttrib< TinyVector< T, D >> &pb) |
| template<typename T , unsigned D> | |
| T | Dot (const ParticleAttrib< TinyVector< std::complex< T >, D >> &pa, const ParticleAttrib< TinyVector< std::complex< T >, D >> &pb) |
| template<typename T , unsigned D> | |
| std::complex< T > | CplxDot (const ParticleAttrib< TinyVector< std::complex< T >, D >> &pa, const ParticleAttrib< TinyVector< std::complex< T >, D >> &pb) |
| template<unsigned D> | |
| double | Dot_CC (const ParticleAttrib< TinyVector< std::complex< double >, D >> &pa, const ParticleAttrib< TinyVector< std::complex< double >, D >> &pb) |
| template<typename T > | |
| T | Sum (const ParticleAttrib< T > &pa) |
| template<typename T > | |
| T | Sum (const ParticleAttrib< std::complex< T >> &pa) |
| template<typename T > | |
| std::complex< T > | CplxSum (const ParticleAttrib< std::complex< T >> &pa) |
| template<class T , unsigned D> | |
| void | Copy (const ParticleAttrib< TinyVector< std::complex< T >, D >> &c, ParticleAttrib< TinyVector< T, D >> &r) |
| template<class T , unsigned D> | |
| void | Copy (const ParticleAttrib< TinyVector< T, D >> &c, ParticleAttrib< TinyVector< T, D >> &r) |
| template<class PL , class PV > | |
| void | convert (const PL &lat, const PV &pin, PV &pout) |
| template<class PL , class PV > | |
| void | convert2Cart (const PL &lat, PV &pin) |
| template<class PL , class PV > | |
| void | convert2Unit (const PL &lat, PV &pin) |
| template<class PL , class PV > | |
| void | wrapAroundBox (const PL &lat, const PV &pin, PV &pout) |
| std::ostream & | operator<< (std::ostream &o_stream, PosUnit pos_unit) |
| write unit type in human readable format More... | |
| std::istream & | operator>> (std::istream &i_stream, PosUnit &pos_unit) |
| Read unit type recorded in int. More... | |
| template<class T , class RG > | |
| void | assignGaussRand (T *restrict a, unsigned n, RG &rng) |
| template<class T , class RG > | |
| void | assignUniformRand (T *restrict a, unsigned n, RG &rng) |
| template<typename T , unsigned D, class RG > | |
| void | makeGaussRandomWithEngine (ParticleAttrib< TinyVector< T, D >> &a, RG &rng) |
| template<typename T , unsigned D, class RG > | |
| void | makeGaussRandomWithEngine (std::vector< TinyVector< T, D >> &a, RG &rng) |
| template<typename T , class RG > | |
| void | makeGaussRandomWithEngine (std::vector< T > &a, RG &rng) |
| template<typename T , class RG > | |
| void | makeGaussRandomWithEngine (ParticleAttrib< T > &a, RG &rng) |
| template<CoordsType CT, class RG > | |
| void | makeGaussRandomWithEngine (MCCoords< CT > &a, RG &rng) |
| template<typename T , unsigned D> | |
| void | makeGaussRandom (std::vector< TinyVector< T, D >> &a) |
| template<typename T , unsigned D> | |
| void | makeGaussRandom (Matrix< TinyVector< T, D >> &a) |
| specialized functions: stick to overloading More... | |
| template<typename T , unsigned D> | |
| void | makeGaussRandom (ParticleAttrib< TinyVector< T, D >> &a) |
| template<typename T > | |
| void | makeGaussRandom (ParticleAttrib< T > &a) |
| template<typename T , unsigned D> | |
| void | makeUniformRandom (ParticleAttrib< TinyVector< T, D >> &a) |
| template<typename T > | |
| void | makeUniformRandom (ParticleAttrib< T > &a) |
| template<typename T > | |
| void | makeSphereRandom (ParticleAttrib< TinyVector< T, 3 >> &a) |
| template<typename T > | |
| void | makeSphereRandom (ParticleAttrib< TinyVector< T, 2 >> &a) |
| template<unsigned int D> | |
| void | double_test_case () |
| TEST_CASE ("particle_attrib_ops_double", "[particle_base]") | |
| template<unsigned int D> | |
| void | complex_test_case () |
| TEST_CASE ("particle_attrib_ops_complex", "[particle_base]") | |
| TEST_CASE ("particle_attrib_scalar", "[particle_base]") | |
| TEST_CASE ("particle_attrib_vector", "[particle_base]") | |
| TEST_CASE ("gaussian random array length 1", "[particle_base]") | |
| TEST_CASE ("gaussian random array length 2", "[particle_base]") | |
| TEST_CASE ("gaussian random array length 3", "[particle_base]") | |
| TEST_CASE ("gaussian random particle attrib array length 1", "[particle_base]") | |
| TEST_CASE ("gaussian random input one", "[particle_base]") | |
| TEST_CASE ("gaussian random input zero", "[particle_base]") | |
| std::vector< double > | gauss_random_vals (size_test *3+(size_test *3) % 2+size_test) |
| makeGaussRandomWithEngine (gauss_random_vals, rng) | |
| makeGaussRandomWithEngine (mc_coords_rs, rng) | |
| checkRs (mc_coords_rs.positions) | |
| makeGaussRandomWithEngine (mc_coords_rsspins, rng) | |
| checkRs (mc_coords_rsspins.positions) | |
| for (int i=0;i< size_test;++i) CHECK(Approx(gauss_random_vals[offset_for_rs+i]) | |
| TEST_CASE ("read_lattice_xml", "[particle_io][xml]") | |
| TEST_CASE ("read_lattice_xml_lrhandle", "[particle_io][xml]") | |
| TEST_CASE ("read_particleset_xml", "[particle_io][xml]") | |
| TEST_CASE ("read_particleset_recorder_xml", "[particle_io][xml]") | |
| TEST_CASE ("read_dynamic_spin_eset_xml", "[particle_io][xml]") | |
| TEST_CASE ("read_particle_mass_same_xml", "[particle_io][xml]") | |
| void | setSpeciesProperty (SpeciesSet &tspecies, int sid, xmlNodePtr cur) |
| set the property of a SpeciesSet More... | |
| static const TimerNameList_t< PSetTimers > | generatePSetTimerNames (std::string &obj_name) |
| template void | ParticleSet::mw_makeMove< CoordsType::POS > (const RefVectorWithLeader< ParticleSet > &p_list, Index_t iat, const MCCoords< CoordsType::POS > &displs) |
| template void | ParticleSet::mw_makeMove< CoordsType::POS_SPIN > (const RefVectorWithLeader< ParticleSet > &p_list, Index_t iat, const MCCoords< CoordsType::POS_SPIN > &displs) |
| template void | ParticleSet::mw_accept_rejectMove< CoordsType::POS > (const RefVectorWithLeader< ParticleSet > &p_list, Index_t iat, const std::vector< bool > &isAccepted, bool forward_mode) |
| template void | ParticleSet::mw_accept_rejectMove< CoordsType::POS_SPIN > (const RefVectorWithLeader< ParticleSet > &p_list, Index_t iat, const std::vector< bool > &isAccepted, bool forward_mode) |
| template void | PSdispatcher::flex_makeMove< CoordsType::POS > (const RefVectorWithLeader< ParticleSet > &p_list, int iat, const MCCoords< CoordsType::POS > &displs) const |
| template void | PSdispatcher::flex_makeMove< CoordsType::POS_SPIN > (const RefVectorWithLeader< ParticleSet > &p_list, int iat, const MCCoords< CoordsType::POS_SPIN > &displs) const |
| template void | PSdispatcher::flex_accept_rejectMove< CoordsType::POS > (const RefVectorWithLeader< ParticleSet > &p_list, int iat, const std::vector< bool > &isAccepted, bool forward_mode) const |
| template void | PSdispatcher::flex_accept_rejectMove< CoordsType::POS_SPIN > (const RefVectorWithLeader< ParticleSet > &p_list, int iat, const std::vector< bool > &isAccepted, bool forward_mode) const |
| TEST_CASE ("distance_open_z", "[distance_table][xml]") | |
| TEST_CASE ("distance_open_xy", "[distance_table][xml]") | |
| TEST_CASE ("distance_open_species_deviation", "[distance_table][xml]") | |
| SimulationCell | parse_pbc_fcc_lattice () |
| SimulationCell | parse_pbc_lattice () |
| void | parse_electron_ion_pbc_z (ParticleSet &ions, ParticleSet &electrons) |
| TEST_CASE ("distance_pbc_z", "[distance_table][xml]") | |
| void | test_distance_pbc_z_batched_APIs (DynamicCoordinateKind test_kind) |
| void | test_distance_fcc_pbc_z_batched_APIs (DynamicCoordinateKind test_kind) |
| TEST_CASE ("distance_pbc_z batched APIs", "[distance_table][xml]") | |
| void | test_distance_pbc_z_batched_APIs_ee_NEED_TEMP_DATA_ON_HOST (DynamicCoordinateKind test_kind) |
| TEST_CASE ("distance_pbc_z batched APIs ee NEED_TEMP_DATA_ON_HOST", "[distance_table][xml]") | |
| TEST_CASE ("test_distance_pbc_diamond", "[distance_table][xml]") | |
| TEST_CASE ("DTModes", "[particle]") | |
| TEST_CASE ("MCCoords", "[Particle]") | |
| TEST_CASE ("ParticleSetPool", "[qmcapp]") | |
| TEST_CASE ("ParticleSetPool random", "[qmcapp]") | |
| TEST_CASE ("ParticleSetPool putLattice", "[qmcapp]") | |
| TEST_CASE ("ParticleSet distance table management", "[particle]") | |
| TEST_CASE ("symmetric_distance_table OpenBC", "[particle]") | |
| TEST_CASE ("symmetric_distance_table PBC", "[particle]") | |
| TEST_CASE ("particle set lattice with vacuum", "[particle]") | |
| TEST_CASE ("SampleStack", "[particle]") | |
| TEST_CASE ("SoaDistanceTableAA compute_size", "[distance_table]") | |
| TEST_CASE ("VirtualParticleSet", "[particle]") | |
| TEST_CASE ("walker", "[particle]") | |
| TEST_CASE ("walker assumptions", "[particle]") | |
| Currently significant amounts of code assumes that the Walker by default has "Properties" ending with the LOCALPOTENTIAL element. More... | |
| TEST_CASE ("walker HDF read and write", "[particle]") | |
| TEST_CASE ("walker buffer add, update, restore", "[particle]") | |
| template<class RealType , class PA > | |
| std::ostream & | operator<< (std::ostream &out, const Walker< RealType, PA > &rhs) |
| std::string | make_bandinfo_filename (const std::string &root, int spin, int twist, const Tensor< int, 3 > &tilematrix, int gid) |
| std::string | make_bandgroup_name (const std::string &root, int spin, int twist, const Tensor< int, 3 > &tilematrix, int first, int last) |
| template<typename T > | |
| T | SymTrace (T h00, T h01, T h02, T h11, T h12, T h22, const T gg[6]) |
| compute Trace(H*G) More... | |
| template<typename T > | |
| T | v_m_v (T h00, T h01, T h02, T h11, T h12, T h22, T g1x, T g1y, T g1z, T g2x, T g2y, T g2z) |
| compute vector[3]^T x matrix[3][3] x vector[3] More... | |
| template<typename T > | |
| T | t3_contract (T h000, T h001, T h002, T h011, T h012, T h022, T h111, T h112, T h122, T h222, T g1x, T g1y, T g1z, T g2x, T g2y, T g2z, T g3x, T g3y, T g3z) |
| Coordinate transform for a 3rd rank symmetric tensor representing coordinate derivatives (hence t3_contract, for contraction with vectors). More... | |
| template<typename ST > | |
| std::unique_ptr< BsplineReader > | createBsplineComplex (EinsplineSetBuilder *e, bool hybrid_rep, const std::string &useGPU) |
| create a reader which handles complex (double size real) splines, C2R or C2C case spline storage and computation precision is ST More... | |
| std::unique_ptr< BsplineReader > | createBsplineComplex (EinsplineSetBuilder *e, bool use_single, bool hybrid_rep, const std::string &useGPU) |
| create a reader which handles complex (double size real) splines, C2R or C2C case spline storage and computation precision is double More... | |
| std::unique_ptr< BsplineReader > | createBsplineReal (EinsplineSetBuilder *e, bool use_single, bool hybrid_rep, const std::string &useGPU) |
| create a reader which handles real splines, R2R case spline storage and computation precision is double More... | |
| template<typename ST > | |
| std::unique_ptr< BsplineReader > | createBsplineReal (EinsplineSetBuilder *e, bool hybrid_rep, const std::string &useGPU) |
| create a reader which handles real splines, R2R case spline storage and computation precision is ST More... | |
| template<typename T > | |
| void | unpack4fftw (const Vector< std::complex< T >> &cG, const std::vector< TinyVector< int, 3 >> &gvecs, const TinyVector< int, 3 > &maxg, Array< std::complex< T >, 3 > &fftbox) |
| unpack packed cG to fftbox More... | |
| template<typename T , typename T1 , typename T2 > | |
| void | fix_phase_rotate_c2r (Array< std::complex< T >, 3 > &in, Array< T1, 3 > &out, const TinyVector< T2, 3 > &twist, T &phase_r, T &phase_i) |
| rotate the state after 3dfft More... | |
| template<typename T , typename T1 , typename T2 > | |
| void | fix_phase_rotate_c2c (const Array< std::complex< T >, 3 > &in, Array< std::complex< T1 >, 3 > &out, const TinyVector< T2, 3 > &twist) |
| template<typename T , typename T1 , typename T2 > | |
| void | fix_phase_rotate_c2c (const Array< std::complex< T >, 3 > &in, Array< T1, 3 > &out_r, Array< T1, 3 > &out_i, const TinyVector< T2, 3 > &twist, T &phase_r, T &phase_i) |
| template<typename T , typename T1 > | |
| void | split_real_components_c2c (const Array< std::complex< T >, 3 > &in, Array< T1, 3 > &out_r, Array< T1, 3 > &out_i) |
| Split FFTs into real/imaginary components. More... | |
| template<typename T > | |
| T | compute_norm (const Vector< std::complex< T >> &cG) |
| Compute the norm of an orbital. More... | |
| template<typename T , typename T2 > | |
| void | compute_phase (const Array< std::complex< T >, 3 > &in, const TinyVector< T2, 3 > &twist, T &phase_r, T &phase_i) |
| Compute the phase factor for rotation. More... | |
| template<typename T > | |
| void | fix_phase_rotate (const Array< std::complex< T >, 3 > &e2pi, Array< std::complex< T >, 3 > &in, Array< T, 3 > &out) |
| rotate the state after 3dfft More... | |
| template<typename T > | |
| TinyVector< T, 3 > | IntPart (const TinyVector< T, 3 > &twist) |
| template<typename T > | |
| TinyVector< T, 3 > | FracPart (const TinyVector< T, 3 > &twist) |
| bool | sortByIndex (BandInfo leftB, BandInfo rightB) |
| template<typename T > | |
| sycl::event | applyW_stageV_sycl (sycl::queue &aq, const int *restrict delay_list_gpu, const int delay_count, T *restrict temp_gpu, const int numorbs, const int ndelay, T *restrict V_gpu, const T *restrict Ainv, const std::vector< sycl::event > &dependencies) |
| template sycl::event | applyW_stageV_sycl (sycl::queue &aq, const int *restrict delay_list_gpu, const int delay_count, float *restrict temp_gpu, const int numorbs, const int ndelay, float *restrict V_gpu, const float *restrict Ainv, const std::vector< sycl::event > &dependencies) |
| template sycl::event | applyW_stageV_sycl (sycl::queue &aq, const int *restrict delay_list_gpu, const int delay_count, double *restrict temp_gpu, const int numorbs, const int ndelay, double *restrict V_gpu, const double *restrict Ainv, const std::vector< sycl::event > &dependencies) |
| template sycl::event | applyW_stageV_sycl (sycl::queue &aq, const int *restrict delay_list_gpu, const int delay_count, std::complex< float > *restrict temp_gpu, const int numorbs, const int ndelay, std::complex< float > *restrict V_gpu, const std::complex< float > *restrict Ainv, const std::vector< sycl::event > &dependencies) |
| template sycl::event | applyW_stageV_sycl (sycl::queue &aq, const int *restrict delay_list_gpu, const int delay_count, std::complex< double > *restrict temp_gpu, const int numorbs, const int ndelay, std::complex< double > *restrict V_gpu, const std::complex< double > *restrict Ainv, const std::vector< sycl::event > &dependencies) |
| template<typename T , typename TMAT , typename INDEX > | |
| std::complex< T > | computeLogDet_sycl (sycl::queue &aq, int n, int lda, const TMAT *restrict a, const INDEX *restrict pivot, const std::vector< sycl::event > &dependencies) |
| template std::complex< double > | computeLogDet_sycl (sycl::queue &aq, int n, int lda, const double *restrict a, const std::int64_t *restrict pivot, const std::vector< sycl::event > &dependencies) |
| template std::complex< double > | computeLogDet_sycl (sycl::queue &aq, int n, int lda, const std::complex< double > *restrict a, const std::int64_t *restrict pivot, const std::vector< sycl::event > &dependencies) |
| template<typename T > | |
| sycl::event | applyW_stageV_sycl (sycl::queue &aq, const int *delay_list_gpu, const int delay_count, T *temp_gpu, const int numorbs, const int ndelay, T *V_gpu, const T *Ainv, const std::vector< sycl::event > &dependencies={}) |
| template<typename T , typename TMAT , typename INDEX > | |
| std::complex< T > | computeLogDet_sycl (sycl::queue &aq, int n, int lda, const TMAT *a, const INDEX *pivot, const std::vector< sycl::event > &dependencies={}) |
| std::ostream & | operator<< (std::ostream &out, const ci_configuration &c) |
| std::ostream & | operator<< (std::ostream &out, const ci_configuration2 &c) |
| int | Xgetrf (int n, int m, float *restrict a, int lda, int *restrict piv) |
| wrappers around xgetrf lapack routines More... | |
| int | Xgetrf (int n, int m, std::complex< float > *restrict a, int lda, int *restrict piv) |
| int | Xgetrf (int n, int m, double *restrict a, int lda, int *restrict piv) |
| int | Xgetrf (int n, int m, std::complex< double > *restrict a, int lda, int *restrict piv) |
| int | Xgetri (int n, float *restrict a, int lda, int *restrict piv, float *restrict work, int &lwork) |
| inversion of a float matrix after lu factorization More... | |
| int | Xgetri (int n, std::complex< float > *restrict a, int lda, int *restrict piv, std::complex< float > *restrict work, int &lwork) |
| int | Xgetri (int n, double *restrict a, int lda, int *restrict piv, double *restrict work, int &lwork) |
| int | Xgetri (int n, std::complex< double > *restrict a, int lda, int *restrict piv, std::complex< double > *restrict work, int &lwork) |
| inversion of a std::complex<double> matrix after lu factorization More... | |
| template<typename T , typename T_FP > | |
| void | computeLogDet (const T *restrict diag, int n, const int *restrict pivot, std::complex< T_FP > &logdet) |
| size_t | flat_idx (const int i, const int a, const int n) |
| template<typename VALUE > | |
| VALUE | calcSmallDeterminant (size_t n, const VALUE *table_matrix, const int *it, const size_t nb_cols) |
| bool | Include (int i, int j, int k) |
| bool | Include (int i, int j) |
| template<class T > | |
| bool | putContent2 (std::vector< T > &a, xmlNodePtr cur) |
| bool | readCuspInfo (const std::string &cuspInfoFile, const std::string &objectName, int OrbitalSetSize, Matrix< CuspCorrectionParameters > &info) |
| Read cusp correction parameters from XML file. More... | |
| void | saveCusp (const std::string &filename, const Matrix< CuspCorrectionParameters > &info, const std::string &id) |
| save cusp correction info to a file. More... | |
| void | broadcastCuspInfo (CuspCorrectionParameters ¶m, Communicate &Comm, int root) |
| Broadcast cusp correction parameters. More... | |
| void | splitPhiEta (int center, const std::vector< bool > &corrCenter, LCAOrbitalSet &phi, LCAOrbitalSet &eta) |
| Divide molecular orbital into atomic S-orbitals on this center (phi), and everything else (eta). More... | |
| void | removeSTypeOrbitals (const std::vector< bool > &corrCenter, LCAOrbitalSet &Phi) |
| Remove S atomic orbitals from all molecular orbitals on all centers. More... | |
| void | computeRadialPhiBar (ParticleSet *targetP, ParticleSet *sourceP, int curOrb_, int curCenter_, SPOSet *Phi, Vector< QMCTraits::RealType > &xgrid, Vector< QMCTraits::RealType > &rad_orb, const CuspCorrectionParameters &data) |
| Compute the radial part of the corrected wavefunction. More... | |
| RealType | getOneIdealLocalEnergy (RealType r, RealType Z, RealType beta0) |
| Ideal local energy at one point. More... | |
| void | getIdealLocalEnergy (const ValueVector &pos, RealType Z, RealType Rc, RealType ELorigAtRc, ValueVector &ELideal) |
| Ideal local energy at a vector of points. More... | |
| void | evalX (RealType valRc, GradType gradRc, ValueType lapRc, RealType Rc, RealType Z, RealType C, RealType valAtZero, RealType eta0, TinyVector< ValueType, 5 > &X) |
| Evaluate various orbital quantities that enter as constraints on the correction. More... | |
| void | X2alpha (const TinyVector< ValueType, 5 > &X, RealType Rc, TinyVector< ValueType, 5 > &alpha) |
| Convert constraints to polynomial parameters. More... | |
| RealType | getZeff (RealType Z, RealType etaAtZero, RealType phiBarAtZero) |
| Effective nuclear charge to keep effective local energy finite at zero. More... | |
| RealType | phiBar (const CuspCorrection &cusp, RealType r, OneMolecularOrbital &phiMO) |
| void | getCurrentLocalEnergy (const ValueVector &pos, RealType Zeff, RealType Rc, RealType originalELatRc, CuspCorrection &cusp, OneMolecularOrbital &phiMO, ValueVector &ELcurr) |
| Compute effective local energy at vector of points. More... | |
| RealType | getOriginalLocalEnergy (const ValueVector &pos, RealType Zeff, RealType Rc, OneMolecularOrbital &phiMO, ValueVector &Elorig) |
| Local energy from uncorrected orbital. More... | |
| RealType | getELchi2 (const ValueVector &ELcurr, const ValueVector &ELideal) |
| Sum of squares difference between the current and ideal local energies This is the objective function to be minimized. More... | |
| RealType | evaluateForPhi0Body (RealType phi0, ValueVector &pos, ValueVector &ELcurr, ValueVector &ELideal, CuspCorrection &cusp, OneMolecularOrbital &phiMO, ValGradLap phiAtRc, RealType etaAtZero, RealType ELorigAtRc, RealType Z) |
| RealType | minimizeForPhiAtZero (CuspCorrection &cusp, OneMolecularOrbital &phiMO, RealType Z, RealType eta0, ValueVector &pos, ValueVector &ELcurr, ValueVector &ELideal, RealType start_phi0) |
| Minimize chi2 with respect to phi at zero for a fixed Rc. More... | |
| void | minimizeForRc (CuspCorrection &cusp, OneMolecularOrbital &phiMO, RealType Z, RealType Rc_init, RealType Rc_max, RealType eta0, ValueVector &pos, ValueVector &ELcurr, ValueVector &ELideal) |
| Minimize chi2 with respect to Rc and phi at zero. More... | |
| void | applyCuspCorrection (const Matrix< CuspCorrectionParameters > &info, ParticleSet &targetPtcl, ParticleSet &sourcePtcl, LCAOrbitalSet &lcao, SoaCuspCorrection &cusp, const std::string &id) |
| void | generateCuspInfo (Matrix< CuspCorrectionParameters > &info, const ParticleSet &targetPtcl, const ParticleSet &sourcePtcl, const LCAOrbitalSet &lcao, const std::string &id, Communicate &Comm) |
| bool | is_same (const xmlChar *a, const char *b) |
| template<typename T , unsigned D, typename Alloc > | |
| void | Product_ABt (const VectorSoaContainer< T, D > &A, const Matrix< T, Alloc > &B, VectorSoaContainer< T, D > &C) |
| Find a better place for other user classes, Matrix should be padded as well. More... | |
| void | print (OptimizableFunctorBase &func, std::ostream &os, double extent=-1.0) |
| evaluates a functor (value and derivative) and dumps the quantities to output More... | |
| std::string | extractCoefficientsID (xmlNodePtr cur) |
| return the id of the first coefficients. If not found, return an emtpy string More... | |
| template<typename T > | |
| std::complex< T > | convertValueToLog (const std::complex< T > &logpsi) |
| evaluate log(psi) as log(|psi|) and phase More... | |
| template<typename T > | |
| std::complex< T > | convertValueToLog (const T logpsi) |
| TEST_CASE ("cuSolverInverter_bench", "[wavefunction][benchmark]") | |
| std::ostream & | operator<< (std::ostream &out, const DiracComputeBenchmarkParameters &dcbmp) |
| TEST_CASE ("DiracMatrixComputeCUDA_large_determinants_benchmark_legacy_1024_4", "[wavefunction][fermion][.benchmark]") | |
| This and other [.benchmark] benchmarks only run if "[benchmark]" is explicitly passed as tag to test. More... | |
| TEST_CASE ("benchmark_DiracMatrixComputeCUDA_vs_legacy_256_10", "[wavefunction][fermion][benchmark]") | |
| This test will run by default. More... | |
| TEST_CASE ("benchmark_DiracMatrixComputeCUDASingle_vs_legacy_256_10", "[wavefunction][fermion][.benchmark]") | |
| Only runs if [benchmark] tag is passed. More... | |
| TEST_CASE ("benchmark_DiracMatrixComputeCUDASingle_vs_legacy_1024_4", "[wavefunction][fermion][.benchmark]") | |
| Only runs if [benchmark] tag is passed. More... | |
| TEST_CASE ("rocSolverInverter_bench", "[wavefunction][benchmark]") | |
| template<typename T > | |
| void | fillIdentityMatrix (Matrix< T > &m) |
| TEST_CASE ("Jastrow 2D", "[wavefunction]") | |
| void | test_cartesian_ao () |
| void | test_dirac_ao () |
| TEST_CASE ("Cartesian Gaussian Ordering", "[wavefunction]") | |
| TEST_CASE ("Dirac Cartesian Gaussian Ordering", "[wavefunction]") | |
| TEST_CASE ("ci_configuration2", "[wavefunction]") | |
| TEST_CASE ("CompositeSPO::diamond_1x1x1", "[wavefunction") | |
| TEST_CASE ("ConstantSPOSet", "[wavefunction]") | |
| TEST_CASE ("Gaussian Functor", "[wavefunction]") | |
| TEST_CASE ("CountingJastrow", "[wavefunction]") | |
| TEST_CASE ("cuBLAS_LU::computeLogDet", "[wavefunction][CUDA]") | |
| Single double computeLogDet. More... | |
| TEST_CASE ("cuBLAS_LU::computeLogDet_complex", "[wavefunction][CUDA]") | |
| TEST_CASE ("cuBLAS_LU::computeLogDet_float", "[wavefunction][CUDA]") | |
| while this working is a good test, in production code its likely we want to widen the matrix M to double and thereby the LU matrix as well. More... | |
| std::vector< StdComp, CUDAHostAllocator< StdComp > > | dev_lu (lu.size()) |
| std::vector< StdComp, CUDAHostAllocator< StdComp > > | dev_lu2 (lu2.size()) |
| std::vector< StdComp *, CUDAHostAllocator< StdComp * > > | lus (batch_size) |
| std::vector< StdComp *, CUDAAllocator< StdComp * > > | dev_lus (batch_size) |
| std::vector< StdComp, CUDAHostAllocator< StdComp > > | log_values (batch_size) |
| std::vector< StdComp, CUDAAllocator< StdComp > > | dev_log_values (batch_size) |
| std::vector< int, CUDAAllocator< int > > | dev_pivots (pivots.size()) |
| cudaErrorCheck (cudaMemcpyAsync(dev_lu.data(), lu.data(), sizeof(decltype(lu)::value_type) *lu.size(), cudaMemcpyHostToDevice, hstream), "cudaMemcpyAsync failed copying log_values to device") | |
| cudaErrorCheck (cudaMemcpyAsync(dev_lu2.data(), lu2.data(), sizeof(decltype(lu2)::value_type) *lu2.size(), cudaMemcpyHostToDevice, hstream), "cudaMemcpyAsync failed copying log_values to device") | |
| cudaErrorCheck (cudaMemcpyAsync(dev_lus.data(), lus.data(), sizeof(decltype(lus)::value_type) *lus.size(), cudaMemcpyHostToDevice, hstream), "cudaMemcpyAsync failed copying log_values to device") | |
| cudaErrorCheck (cudaMemcpyAsync(dev_pivots.data(), pivots.data(), sizeof(int) *pivots.size(), cudaMemcpyHostToDevice, hstream), "cudaMemcpyAsync failed copying log_values to device") | |
| cudaErrorCheck (cudaMemcpyAsync(log_values.data(), dev_log_values.data(), sizeof(std::complex< double >) *2, cudaMemcpyDeviceToHost, hstream), "cudaMemcpyAsync failed copying log_values from device") | |
| cudaErrorCheck (cudaStreamSynchronize(hstream), "cudaStreamSynchronize failed!") | |
| CHECK (log_values[0]==ComplexApprox(std::complex< double >{ 5.603777579195571, -6.1586603331188225 })) | |
| CHECK (log_values[1]==ComplexApprox(std::complex< double >{ 5.531331998282581, -8.805487075984523 })) | |
| TEST_CASE ("cuBLAS_LU::getrf_batched_complex", "[wavefunction][CUDA]") | |
| std::vector< double, CUDAAllocator< double > > | devM_vec (M_vec.size()) |
| std::vector< double, CUDAAllocator< double > > | devM2_vec (M2_vec.size()) |
| std::vector< double *, CUDAAllocator< double * > > | devMs (Ms.size()) |
| std::vector< int, CUDAHostAllocator< int > > | pivots (8, -1.0) |
| std::vector< int, CUDAHostAllocator< int > > | infos (8, 1.0) |
| std::vector< int, CUDAAllocator< int > > | dev_infos (pivots.size()) |
| cudaErrorCheck (cudaMemcpyAsync(devM_vec.data(), M_vec.data(), sizeof(decltype(M_vec)::value_type) *M_vec.size(), cudaMemcpyHostToDevice, hstream), "cudaMemcpyAsync failed copying M to device") | |
| cudaErrorCheck (cudaMemcpyAsync(devM2_vec.data(), M2_vec.data(), sizeof(decltype(M2_vec)::value_type) *M2_vec.size(), cudaMemcpyHostToDevice, hstream), "cudaMemcpyAsync failed copying M2 to device") | |
| cudaErrorCheck (cudaMemcpyAsync(devMs.data(), Ms.data(), sizeof(decltype(Ms)::value_type) *Ms.size(), cudaMemcpyHostToDevice, hstream), "cudaMemcpyAsync failed copying Ms to device") | |
| cudaErrorCheck (cudaMemcpyAsync(M_vec.data(), devM_vec.data(), sizeof(decltype(M_vec)::value_type) *M_vec.size(), cudaMemcpyDeviceToHost, hstream), "cudaMemcpyAsync failed copying invM from device") | |
| cudaErrorCheck (cudaMemcpyAsync(M2_vec.data(), devM2_vec.data(), sizeof(decltype(M2_vec)::value_type) *M2_vec.size(), cudaMemcpyDeviceToHost, hstream), "cudaMemcpyAsync failed copying invM from device") | |
| cudaErrorCheck (cudaMemcpyAsync(pivots.data(), dev_pivots.data(), sizeof(int) *pivots.size(), cudaMemcpyDeviceToHost, hstream), "cudaMemcpyAsync failed copying pivots from device") | |
| testing::MatrixAccessor< double > | M_mat (M_vec.data(), 4, 4) |
| testing::MatrixAccessor< double > | lu_mat (lu.data(), 4, 4) |
| testing::MatrixAccessor< double > | M2_mat (M2_vec.data(), 4, 4) |
| testing::MatrixAccessor< double > | lu2_mat (lu2.data(), 4, 4) |
| checkArray (real_pivot, pivots, 8) | |
| CHECKED_ELSE (check_matrix_result.result) | |
| TEST_CASE ("cuBLAS_LU::getri_batched", "[wavefunction][CUDA]") | |
| TEMPLATE_TEST_CASE ("cuSolverInverter", "[wavefunction][fermion]", double, float) | |
| template<typename T1 , typename T2 > | |
| void | check_matrix (Matrix< T1 > &a, Matrix< T2 > &b) |
| template<typename DET > | |
| void | test_DiracDeterminant_first (const DetMatInvertor inverter_kind) |
| TEST_CASE ("DiracDeterminant_first", "[wavefunction][fermion]") | |
| template<typename DET > | |
| void | test_DiracDeterminant_second (const DetMatInvertor inverter_kind) |
| TEST_CASE ("DiracDeterminant_second", "[wavefunction][fermion]") | |
| template<typename DET > | |
| void | test_DiracDeterminant_delayed_update (const DetMatInvertor inverter_kind) |
| TEST_CASE ("DiracDeterminant_delayed_update", "[wavefunction][fermion]") | |
| template<PlatformKind PL> | |
| void | test_DiracDeterminantBatched_first () |
| TEST_CASE ("DiracDeterminantBatched_first", "[wavefunction][fermion]") | |
| template<PlatformKind PL> | |
| void | test_DiracDeterminantBatched_second () |
| TEST_CASE ("DiracDeterminantBatched_second", "[wavefunction][fermion]") | |
| template<PlatformKind PL> | |
| void | test_DiracDeterminantBatched_delayed_update (int delay_rank, DetMatInvertor matrix_inverter_kind) |
| TEST_CASE ("DiracDeterminantBatched_delayed_update", "[wavefunction][fermion]") | |
| TEST_CASE ("DiracMatrix_identity", "[wavefunction][fermion]") | |
| TEST_CASE ("DiracMatrix_inverse", "[wavefunction][fermion]") | |
| TEST_CASE ("DiracMatrix_inverse_matching", "[wavefunction][fermion]") | |
| TEST_CASE ("DiracMatrix_inverse_matching_2", "[wavefunction][fermion]") | |
| TEST_CASE ("DiracMatrix_inverse_complex", "[wavefunction][fermion]") | |
| This test case is meant to match the cuBLAS_LU::getrf_batched_complex. More... | |
| TEST_CASE ("DiracMatrix_update_row", "[wavefunction][fermion]") | |
| TEST_CASE ("DiracMatrixComputeCUDA_cuBLAS_geam_call", "[wavefunction][fermion]") | |
| TEST_CASE ("DiracMatrixComputeCUDA_different_batch_sizes", "[wavefunction][fermion]") | |
| TEST_CASE ("DiracMatrixComputeCUDA_complex_determinants_against_legacy", "[wavefunction][fermion]") | |
| TEST_CASE ("DiracMatrixComputeCUDA_large_determinants_against_legacy", "[wavefunction][fermion]") | |
| TEST_CASE ("DiracMatrixComputeOMPTarget_different_batch_sizes", "[wavefunction][fermion]") | |
| TEST_CASE ("DiracMatrixComputeOMPTarget_large_determinants_against_legacy", "[wavefunction][fermion]") | |
| TEST_CASE ("Einspline SPO from HDF diamond_1x1x1", "[wavefunction]") | |
| TEST_CASE ("Einspline SPO from HDF diamond_2x1x1 5 electrons", "[wavefunction]") | |
| TEST_CASE ("EinsplineSetBuilder CheckLattice", "[wavefunction]") | |
| TEST_CASE ("Einspline SPO from HDF NiO a16 97 electrons", "[wavefunction]") | |
| TEST_CASE ("ExampleHe", "[wavefunction]") | |
| TEST_CASE ("Hybridrep SPO from HDF diamond_1x1x1", "[wavefunction]") | |
| TEST_CASE ("Hybridrep SPO from HDF diamond_2x1x1", "[wavefunction]") | |
| TEST_CASE ("BSpline functor zero", "[wavefunction]") | |
| TEST_CASE ("BSpline functor one", "[wavefunction]") | |
| void | test_J1_spline (const DynamicCoordinateKind kind_selected) |
| TEST_CASE ("BSpline builder Jastrow J1", "[wavefunction]") | |
| TEST_CASE ("J1 evaluate derivatives Jastrow", "[wavefunction]") | |
| TEST_CASE ("J1 evaluate derivatives Jastrow with two species", "[wavefunction]") | |
| TEST_CASE ("J1 evaluate derivatives Jastrow with two species one without Jastrow", "[wavefunction]") | |
| TEST_CASE ("J1 spin evaluate derivatives Jastrow", "[wavefunction]") | |
| TEST_CASE ("J1 spin evaluate derivatives multiparticle Jastrow", "[wavefunction]") | |
| TEST_CASE ("BSpline builder Jastrow J2", "[wavefunction]") | |
| TEST_CASE ("TwoBodyJastrow simple", "[wavefunction]") | |
| TEST_CASE ("TwoBodyJastrow one species and two variables", "[wavefunction]") | |
| ParticleSet | get_two_species_particleset (const SimulationCell &simulation_cell) |
| TEST_CASE ("TwoBodyJastrow two variables", "[wavefunction]") | |
| TEST_CASE ("TwoBodyJastrow variables fail", "[wavefunction]") | |
| TEST_CASE ("TwoBodyJastrow other variables", "[wavefunction]") | |
| TEST_CASE ("TwoBodyJastrow Jastrow three particles of three types", "[wavefunction]") | |
| TEST_CASE ("kspace jastrow", "[wavefunction]") | |
| TEST_CASE ("lattice gaussian", "[wavefunction]") | |
| void | test_LCAO_DiamondC_2x1x1_real (const bool useOffload) |
| void | test_LCAO_DiamondC_2x1x1_cplx (const bool useOffload) |
| TEST_CASE ("LCAOrbitalSet batched PBC DiamondC", "[wavefunction]") | |
| TEST_CASE ("LCAOrbitalSet batched PBC DiamondC offload", "[wavefunction]") | |
| TEST_CASE ("LCAOrbitalBuilder", "[wavefunction][LCAO]") | |
| void | test_He (bool transform) |
| TEST_CASE ("ReadMolecularOrbital GTO He", "[wavefunction]") | |
| TEST_CASE ("ReadMolecularOrbital Numerical He", "[wavefunction]") | |
| void | test_He_mw (bool transform) |
| TEST_CASE ("mw_evaluate Numerical He", "[wavefunction]") | |
| void | test_EtOH_mw (bool transform) |
| TEST_CASE ("mw_evaluate Numerical EtOH", "[wavefunction]") | |
| TEST_CASE ("mw_evaluate GTO EtOH", "[wavefunction]") | |
| void | test_Ne (bool transform) |
| TEST_CASE ("ReadMolecularOrbital GTO Ne", "[wavefunction]") | |
| TEST_CASE ("ReadMolecularOrbital Numerical Ne", "[wavefunction]") | |
| TEST_CASE ("ReadMolecularOrbital HCN", "[wavefunction]") | |
| void | test_HCN (bool transform) |
| TEST_CASE ("ReadMolecularOrbital GTO HCN", "[wavefunction]") | |
| TEST_CASE ("ReadMolecularOrbital Numerical HCN", "[wavefunction]") | |
| void | test_lcao_spinor () |
| void | test_lcao_spinor_excited () |
| void | test_lcao_spinor_ion_derivs () |
| TEST_CASE ("ReadMolecularOrbital GTO spinor", "[wavefunction]") | |
| TEST_CASE ("ReadMolecularOrbital GTO spinor with excited", "[wavefunction]") | |
| TEST_CASE ("spinor ion derivatives for molecule", "[wavefunction]") | |
| TEST_CASE ("SmallMatrixDetCalculator::evaluate-Small", "[wavefunction][fermion][multidet]") | |
| Simple synthetic test case will trip on changes in this method. More... | |
| void | test_LiH_msd (const std::string &spo_xml_string, const std::string &check_sponame, int check_spo_size, int check_basisset_size, int test_nlpp_algorithm_batched, int test_batched_api) |
| TEST_CASE ("LiH multi Slater dets table_method", "[wavefunction]") | |
| TEST_CASE ("LiH multi Slater dets precomputed_table_method", "[wavefunction]") | |
| TEST_CASE ("LogGridLight", "[wavefunction][LCAO]") | |
| TEST_CASE ("MultiQuinticSpline", "[wavefunction][LCAO]") | |
| TEST_CASE ("NaNguard", "[wavefunction]") | |
| TEST_CASE ("Test OptimizableObject", "[wavefunction]") | |
| TEST_CASE ("OptimizableObject HDF output and input", "[wavefunction]") | |
| TEST_CASE ("Pade functor", "[wavefunction]") | |
| TEST_CASE ("Pade2 functor", "[wavefunction]") | |
| TEST_CASE ("Pade Jastrow", "[wavefunction]") | |
| TEST_CASE ("Pade2 Jastrow", "[wavefunction]") | |
| TEST_CASE ("PolynomialFunctor3D functor zero", "[wavefunction]") | |
| void | test_J3_polynomial3D (const DynamicCoordinateKind kind_selected) |
| TEST_CASE ("PolynomialFunctor3D Jastrow", "[wavefunction]") | |
| TEST_CASE ("PlaneWave SPO from HDF for BCC H", "[wavefunction]") | |
| TEST_CASE ("PlaneWave SPO from HDF for LiH arb", "[wavefunction]") | |
| void | test_C_diamond () |
| TEST_CASE ("ReadMolecularOrbital GTO Carbon Diamond", "[wavefunction]") | |
| TEMPLATE_TEST_CASE ("rocSolverInverter", "[wavefunction][fermion]", double, float) | |
| TEST_CASE ("RotatedSPOs via SplineR2R", "[wavefunction]") | |
| TEST_CASE ("RotatedSPOs createRotationIndices", "[wavefunction]") | |
| TEST_CASE ("RotatedSPOs constructAntiSymmetricMatrix", "[wavefunction]") | |
| TEST_CASE ("RotatedSPOs exponentiate matrix", "[wavefunction]") | |
| TEST_CASE ("RotatedSPOs log matrix", "[wavefunction]") | |
| TEST_CASE ("RotatedSPOs exp-log matrix", "[wavefunction]") | |
| TEST_CASE ("RotatedSPOs hcpBe", "[wavefunction]") | |
| TEST_CASE ("RotatedSPOs construct delta matrix", "[wavefunction]") | |
| TEST_CASE ("RotatedSPOs read and write parameters", "[wavefunction]") | |
| TEST_CASE ("RotatedSPOs read and write parameters history", "[wavefunction]") | |
| TEST_CASE ("RotatedSPOs mw_ APIs", "[wavefunction]") | |
| void | setupParticleSetPool (ParticleSetPool &pp) |
| void | setupParticleSetPoolBe (ParticleSetPool &pp) |
| std::string | setupRotationXML (const std::string &rot_angle_up, const std::string &rot_angle_down, const std::string &coeff_up, const std::string &coeff_down) |
| TEST_CASE ("Rotated LCAO WF0 zero angle", "[qmcapp]") | |
| TEST_CASE ("Rotated LCAO WF1", "[qmcapp]") | |
| TEST_CASE ("Rotated LCAO WF2 with jastrow", "[qmcapp]") | |
| TEST_CASE ("Rotated LCAO WF1, MO coeff rotated, zero angle", "[qmcapp]") | |
| TEST_CASE ("Rotated LCAO WF1 MO coeff rotated, half angle", "[qmcapp]") | |
| TEST_CASE ("Rotated LCAO rotation consistency", "[qmcapp]") | |
| TEST_CASE ("Rotated LCAO Be single determinant", "[qmcapp]") | |
| TEST_CASE ("Rotated LCAO Be multi determinant with one determinant", "[qmcapp]") | |
| TEST_CASE ("Rotated LCAO Be two determinant", "[qmcapp]") | |
| TEST_CASE ("RPA Jastrow", "[wavefunction]") | |
| TEST_CASE ("ShortRangeCuspJastrowFunctor", "[wavefunction]") | |
| TEST_CASE ("SlaterDet mw_ APIs", "[wavefunction]") | |
| TEST_CASE ("readCuspInfo", "[wavefunction]") | |
| TEST_CASE ("applyCuspInfo", "[wavefunction]") | |
| TEST_CASE ("HCN MO with cusp", "[wavefunction]") | |
| TEST_CASE ("Ethanol MO with cusp", "[wavefunction]") | |
| TEST_CASE ("broadcastCuspInfo", "[wavefunction]") | |
| TEST_CASE ("Spline applyRotation zero rotation", "[wavefunction]") | |
| TEST_CASE ("Spline applyRotation one rotation", "[wavefunction]") | |
| TEST_CASE ("Spline applyRotation two rotations", "[wavefunction]") | |
| void | test_He_sto3g_xml_input (const std::string &spo_xml_string) |
| TEST_CASE ("SPO input spline from xml He_sto3g", "[wavefunction]") | |
| void | test_LiH_msd_xml_input (const std::string &spo_xml_string, const std::string &check_sponame, int check_spo_size, int check_basisset_size) |
| TEST_CASE ("SPO input spline from xml LiH_msd", "[wavefunction]") | |
| void | test_LiH_msd_xml_input_with_positron (const std::string &spo_xml_string, const std::string &check_sponame, int check_spo_size, int check_basisset_size) |
| TEST_CASE ("SPO input spline from xml LiH_msd arbitrary species", "[wavefunction]") | |
| TEST_CASE ("SPO input spline from h5 LiH_msd arbitrary species", "[wavefunction]") | |
| void | test_diamond_2x1x1_xml_input (const std::string &spo_xml_string) |
| TEST_CASE ("SPO input spline from HDF diamond_2x1x1", "[wavefunction]") | |
| template<typename T , typename T_FP > | |
| void | test_inverse (const std::int64_t N) |
| TEMPLATE_TEST_CASE ("syclSolverInverter", "[wavefunction][fermion]", double, float) | |
| TEST_CASE ("TrialWaveFunction_diamondC_1x1x1", "[wavefunction]") | |
| template<class DiracDet , class SPO_precision > | |
| void | testTrialWaveFunction_diamondC_2x1x1 (const int ndelay, const OffloadSwitches &offload_switches) |
| Templated test of TrialWF with different DiracDet flavors. More... | |
| TEST_CASE ("TrialWaveFunction_diamondC_2x1x1", "[wavefunction]") | |
| std::unique_ptr< TrialWaveFunction > | setup_He_wavefunction (Communicate *c, ParticleSet &elec, ParticleSet &ions, const WaveFunctionFactory::PSetMap &particle_set_map) |
| TEST_CASE ("TrialWaveFunction flex_evaluateParameterDerivatives", "[wavefunction]") | |
| UPtrVector< ParticleSet::ParticleGradient > | create_particle_gradient (int nelec, int nentry) |
| UPtrVector< ParticleSet::ParticleLaplacian > | create_particle_laplacian (int nelec, int nentry) |
| TEST_CASE ("TrialWaveFunction flex_evaluateDeltaLogSetup", "[wavefunction]") | |
| TEST_CASE ("TWFGrads", "[QMCWaveFunctions]") | |
| TEST_CASE ("UserJastrowFunctor", "[wavefunction]") | |
| TEST_CASE ("WaveFunctionFactory", "[wavefunction]") | |
| TEST_CASE ("WaveFunctionPool", "[qmcapp]") | |
| static TimerNameList_t< TimerEnum > | create_names (std::string_view myName) |
| template void | TrialWaveFunction::mw_evalGrad< CoordsType::POS > (const RefVectorWithLeader< TrialWaveFunction > &wf_list, const RefVectorWithLeader< ParticleSet > &p_list, int iat, TWFGrads< CoordsType::POS > &grads) |
| template void | TrialWaveFunction::mw_evalGrad< CoordsType::POS_SPIN > (const RefVectorWithLeader< TrialWaveFunction > &wf_list, const RefVectorWithLeader< ParticleSet > &p_list, int iat, TWFGrads< CoordsType::POS_SPIN > &grads) |
| template void | TrialWaveFunction::mw_calcRatioGrad< CoordsType::POS > (const RefVectorWithLeader< TrialWaveFunction > &wf_list, const RefVectorWithLeader< ParticleSet > &p_list, int iat, std::vector< PsiValue > &ratios, TWFGrads< CoordsType::POS > &grads) |
| template void | TrialWaveFunction::mw_calcRatioGrad< CoordsType::POS_SPIN > (const RefVectorWithLeader< TrialWaveFunction > &wf_list, const RefVectorWithLeader< ParticleSet > &p_list, int iat, std::vector< PsiValue > &ratios, TWFGrads< CoordsType::POS_SPIN > &grads) |
| template void | TWFdispatcher::flex_evalGrad< CoordsType::POS > (const RefVectorWithLeader< TrialWaveFunction > &wf_list, const RefVectorWithLeader< ParticleSet > &p_list, int iat, TWFGrads< CoordsType::POS > &grads) const |
| template void | TWFdispatcher::flex_evalGrad< CoordsType::POS_SPIN > (const RefVectorWithLeader< TrialWaveFunction > &wf_list, const RefVectorWithLeader< ParticleSet > &p_list, int iat, TWFGrads< CoordsType::POS_SPIN > &grads) const |
| template void | TWFdispatcher::flex_calcRatioGrad< CoordsType::POS > (const RefVectorWithLeader< TrialWaveFunction > &wf_list, const RefVectorWithLeader< ParticleSet > &p_list, int iat, std::vector< PsiValue > &ratios, TWFGrads< CoordsType::POS > &grads) const |
| template void | TWFdispatcher::flex_calcRatioGrad< CoordsType::POS_SPIN > (const RefVectorWithLeader< TrialWaveFunction > &wf_list, const RefVectorWithLeader< ParticleSet > &p_list, int iat, std::vector< PsiValue > &ratios, TWFGrads< CoordsType::POS_SPIN > &grads) const |
| template void | WaveFunctionComponent::mw_evalGrad< CoordsType::POS > (const RefVectorWithLeader< WaveFunctionComponent > &wfc_list, const RefVectorWithLeader< ParticleSet > &p_list, int iat, TWFGrads< CoordsType::POS > &grad_now) const |
| template void | WaveFunctionComponent::mw_evalGrad< CoordsType::POS_SPIN > (const RefVectorWithLeader< WaveFunctionComponent > &wfc_list, const RefVectorWithLeader< ParticleSet > &p_list, int iat, TWFGrads< CoordsType::POS_SPIN > &grad_now) const |
| template void | WaveFunctionComponent::mw_ratioGrad< CoordsType::POS > (const RefVectorWithLeader< WaveFunctionComponent > &wfc_list, const RefVectorWithLeader< ParticleSet > &p_list, int iat, std::vector< PsiValue > &ratios, TWFGrads< CoordsType::POS > &grad_new) const |
| template void | WaveFunctionComponent::mw_ratioGrad< CoordsType::POS_SPIN > (const RefVectorWithLeader< WaveFunctionComponent > &wfc_list, const RefVectorWithLeader< ParticleSet > &p_list, int iat, std::vector< PsiValue > &ratios, TWFGrads< CoordsType::POS_SPIN > &grad_new) const |
| template<typename T , unsigned D> | |
| T | laplacian (const TinyVector< T, D > &g, T l) |
| compute real(laplacian) More... | |
| template<typename T , unsigned D> | |
| T | laplacian (const TinyVector< std::complex< T >, D > &g, const std::complex< T > &l) |
| specialization of laplacian with complex g & l More... | |
| template<typename T , unsigned D> | |
| T | dlaplacian (const TinyVector< T, D > &g, const T l, const TinyVector< T, D > &gg, const T gl, const T ideriv) |
Convenience function to compute 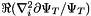 . More... . More... | |
| template<typename T , unsigned D> | |
| T | dlaplacian (const TinyVector< std::complex< T >, D > &g, const std::complex< T > l, const TinyVector< std::complex< T >, D > &gg, const std::complex< T > gl, const std::complex< T > ideriv) |
| RealType | accum_constant (CombinedTraceSample< TraceReal > *etrace, RealType weight=1.0) |
| template<typename T > | |
| void | accum_sample (std::vector< Value_t > &E_samp, CombinedTraceSample< T > *etrace, RealType weight=1.0) |
| template<typename T > | |
| void | accum_sample (std::vector< Value_t > &E_samp, TraceSample< T > *etrace, RealType weight=1.0) |
| double | extrap (int N, TinyVector< double, 3 > g_124) |
| double | extrap (int N, TinyVector< double, 2 > g_12) |
| QMCTraits::TensorType | generateRandomRotationMatrix (RandomBase< QMCTraits::FullPrecRealType > &rng) |
| Create a random 3D rotation matrix using a random generator. More... | |
| TEST_CASE ("Bare Kinetic Energy", "[hamiltonian]") | |
| TEST_CASE ("Bare KE Pulay PBC", "[hamiltonian]") | |
| void | testElecCase (double mass_up, double mass_dn, std::function< void(RefVectorWithLeader< OperatorBase > &o_list, RefVectorWithLeader< TrialWaveFunction > &twf_list, RefVectorWithLeader< ParticleSet > &p_list, Matrix< Real > &kinetic_energies, std::vector< ListenerVector< Real >> &listeners, std::vector< ListenerVector< Real >> &ion_listeners)> tests) |
| Provide a test scope parameterized on electron species mass that then can run a set of tests using its objects. More... | |
| auto | getTestCaseForWeights (Real value1, Real value2, Real value3) |
| just set the values to test the electron weights More... | |
| TEST_CASE ("BareKineticEnergyListener", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A BCC H", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A elec", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A BCC", "[hamiltonian]") | |
| void | test_CoulombPBCAA_3p (DynamicCoordinateKind kind) |
| TEST_CASE ("Coulomb PBC A-A BCC 3 particles", "[hamiltonian]") | |
| TEST_CASE ("CoulombAA::mw_evaluatePerParticle", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A Ewald3D", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A BCC H Ewald3D", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A elec Ewald3D", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-B", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-B BCC H", "[hamiltonian]") | |
| TEST_CASE ("CoulombAB::Listener", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-B Ewald3D", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-B BCC H Ewald3D", "[hamiltonian]") | |
| TEST_CASE ("Density Estimator", "[hamiltonian]") | |
| TEST_CASE ("Density Estimator evaluate exception", "[hamiltonian]") | |
| QMCTraits::RealType | getSplinedSOPot (SOECPComponent *so_comp, int l, double r) |
| TEST_CASE ("ReadFileBuffer_no_file", "[hamiltonian]") | |
| TEST_CASE ("ReadFileBuffer_simple_serial", "[hamiltonian]") | |
| TEST_CASE ("ReadFileBuffer_simple_mpi", "[hamiltonian]") | |
| TEST_CASE ("ReadFileBuffer_ecp", "[hamiltonian]") | |
| TEST_CASE ("ReadFileBuffer_sorep", "[hamiltonian]") | |
| TEST_CASE ("ReadFileBuffer_reopen", "[hamiltonian]") | |
| void | copyGridUnrotatedForTest (NonLocalECPComponent &nlpp) |
| void | copyGridUnrotatedForTest (SOECPComponent &sopp) |
| TEST_CASE ("Evaluate_ecp", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A Ewald2D square", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A Ewald2D body center", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A Ewald2D triangular", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A Ewald2D honeycomb", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A Ewald2D tri. in rect.", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A Ewald Quasi2D exception", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A Ewald Quasi2D square", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A Ewald Quasi2D triangular", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A Ewald Quasi2D staggered square", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A Ewald Quasi2D staggered square 2x2", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A Ewald Quasi2D staggered triangle", "[hamiltonian]") | |
| TEST_CASE ("Coulomb PBC A-A Ewald Quasi2D staggered triangle 2x2", "[hamiltonian]") | |
| TEST_CASE ("EwaldRef", "[hamiltonian]") | |
| TEST_CASE ("Bare Force", "[hamiltonian]") | |
| void | check_force_copy (ForceChiesaPBCAA &force, ForceChiesaPBCAA &force2) |
| TEST_CASE ("Chiesa Force", "[hamiltonian]") | |
| TEST_CASE ("Ceperley Force", "[hamiltonian]") | |
| TEST_CASE ("Ion-ion Force", "[hamiltonian]") | |
| TEST_CASE ("AC Force", "[hamiltonian]") | |
| TEST_CASE ("Chiesa Force BCC H Ewald3D", "[hamiltonian]") | |
| TEST_CASE ("fccz sr lr clone", "[hamiltonian]") | |
| TEST_CASE ("fccz h3", "[hamiltonian]") | |
| std::unique_ptr< ParticleSet > | createElectronParticleSet (const SimulationCell &simulation_cell) |
| TEST_CASE ("HamiltonianFactory", "[hamiltonian]") | |
| TEST_CASE ("HamiltonianFactory pseudopotential", "[hamiltonian]") | |
| TEST_CASE ("HamiltonianPool", "[qmcapp]") | |
| void | create_CN_particlesets (ParticleSet &elec, ParticleSet &ions) |
| void | create_C_pbc_particlesets (ParticleSet &elec, ParticleSet &ions) |
| QMCHamiltonian & | create_CN_Hamiltonian (HamiltonianFactory &hf) |
| TEST_CASE ("Eloc_Derivatives:slater_noj", "[hamiltonian]") | |
| TEST_CASE ("Eloc_Derivatives:slater_wj", "[hamiltonian]") | |
| TEST_CASE ("Eloc_Derivatives:multislater_noj", "[hamiltonian]") | |
| TEST_CASE ("Eloc_Derivatives:multislater_wj", "[hamiltonian]") | |
| TEST_CASE ("Eloc_Derivatives:proto_sd_noj", "[hamiltonian]") | |
| TEST_CASE ("Eloc_Derivatives:proto_sd_wj", "[hamiltonian]") | |
| TEST_CASE ("NonLocalECPotential", "[hamiltonian]") | |
| TEST_CASE ("NonLocalTOperator", "[hamiltonian]") | |
| CHECK (oh.lower_bound==0) | |
| TEST_CASE ("ObservableHelper::set_dimensions", "[hamiltonian]") | |
| hFile | create (filename) |
| oh | addProperty (propertyFloat, "propertyFloat", hFile) |
| oh | addProperty (propertyTensor, "propertyTensor", hFile) |
| oh | addProperty (propertyMatrix, "propertyMatrix", hFile) |
| oh | addProperty (propertyTensor, "propertyTinyVector", hFile) |
| oh | addProperty (propertyVector, "propertyVector", hFile) |
| oh | addProperty (propertyVectorTinyVector, "propertyVectorTinyVector", hFile) |
| hFile | close () |
| REQUIRE (std::filesystem::exists(filename)) | |
| REQUIRE (std::filesystem::remove(filename)) | |
| TEST_CASE ("Pair Correlation", "[hamiltonian]") | |
| TEST_CASE ("Pair Correlation Pair Index", "[hamiltonian]") | |
| TEST_CASE ("QMCHamiltonian::flex_evaluate", "[hamiltonian]") | |
| TEST_CASE ("integrateListeners", "[hamiltonian]") | |
| QMCHamiltonian + Hamiltonians with listeners integration test. More... | |
| void | test_hcpBe_rotation (bool use_single_det, bool use_nlpp_batched) |
| TEST_CASE ("RotatedSPOs SplineR2R hcpBe values", "[wavefunction]") | |
| TEST_CASE ("SkAll", "[hamiltonian]") | |
| void | doSOECPotentialTest (bool use_VPs) |
| TEST_CASE ("SOECPotential", "[hamiltonian]") | |
| TEST_CASE ("SpaceWarp", "[hamiltonian]") | |
| TEST_CASE ("Stress BCC H Ewald3D", "[hamiltonian]") | |
| template bool | InputSection::setIfInInput< qmcplusplus::MagnetizationDensityInput::Integrator > (qmcplusplus::MagnetizationDensityInput::Integrator &var, const std::string &tag) |
| std::ostream & | operator<< (std::ostream &out, const NEReferencePoints &rhs) |
| template bool | InputSection::setIfInInput< qmcplusplus::OneBodyDensityMatricesInput::Integrator > (qmcplusplus::OneBodyDensityMatricesInput::Integrator &var, const std::string &tag) |
| template<typename REAL > | |
| AxisGrid< REAL > | parseGridInput (std::istringstream &grid_input_stream) |
| Parses the one dimensional grid specification format originated for EnergyDensity This has be refactored from QMCHamiltonian/SpaceGrid.cpp. More... | |
| template AxisGrid< double > | parseGridInput< double > (std::istringstream &grid_input_stream) |
| template AxisGrid< float > | parseGridInput< float > (std::istringstream &grid_input_stream) |
| template bool | InputSection::setIfInInput< ReferencePointsInput::Coord > (ReferencePointsInput::Coord &var, const std::string &tag) |
| std::any | makeReferencePointsInput (xmlNodePtr, std::string &value_label) |
| factory function used by InputSection to make reference points Input More... | |
| void | readNameTypeLikeLegacy (InputSection &input_section_, std::string &name, std::string &type) |
| make input class state consistent with legacy ignoring the distinction between the name and type of estimator for scalar estimators. More... | |
| std::any | makeSpaceGridInput (xmlNodePtr, std::string &value_label) |
| factory function for a SpaceGridInput More... | |
| template<typename T > | |
| void | test_real_accumulator_basic () |
| TEST_CASE ("accumulator basic float", "[estimators]") | |
| TEST_CASE ("accumulator basic double", "[estimators]") | |
| template<typename T > | |
| void | test_real_accumulator () |
| TEST_CASE ("accumulator some values float", "[estimators]") | |
| TEST_CASE ("accumulator some values double", "[estimators]") | |
| template<typename T > | |
| void | test_real_accumulator_weights () |
| TEST_CASE ("accumulator with weights float", "[estimators]") | |
| TEST_CASE ("accumulator with weights double", "[estimators]") | |
| TEST_CASE ("EstimatorManagerCrowd::EstimatorManagerCrowd", "[estimators]") | |
| TEST_CASE ("EstimatorManagerCrowd PerParticleHamiltonianLogger integration", "[estimators]") | |
| TEST_CASE ("EstimatorManagerInput::testInserts", "[estimators]") | |
| TEST_CASE ("EstimatorManagerInput::readXML", "[estimators]") | |
| TEST_CASE ("EstimatorManagerInput::moveFromEstimatorInputs", "[estimators]") | |
| TEST_CASE ("EstimatorManagerInput::moveConstructor", "[estimators]") | |
| TEST_CASE ("EstimatorManagerInput::MergeConstructor", "[estimators]") | |
| CHECK (embt.em.getNumEstimators()==0) | |
| EstimatorManagerInput | emi (estimators_doc.getRoot()) |
| CHECK (emi.get_estimator_inputs().size()==2) | |
| CHECK (emn.getNumEstimators()==2) | |
| CHECK (emnta.getMainEstimator().getName()=="LocalEnergyEstimator") | |
| EstimatorManagerInput | emi2 (estimators_doc2.getRoot()) |
| CHECK (emi2.get_estimator_inputs().size()==2) | |
| CHECK (emn2.getNumEstimators()==2) | |
| CHECK (emnta2.getMainEstimator().getName()=="RMCLocalEnergyEstimator") | |
| TEST_CASE ("EstimatorManagerNew::collectMainEstimators", "[estimators]") | |
| TEST_CASE ("EstimatorManagerNew::collectScalarEstimators", "[estimators]") | |
| TEST_CASE ("EstimatorManagerNew adhoc addVector operator", "[estimators]") | |
| TEST_CASE ("InputSection::InputSection", "[estimators]") | |
| TEST_CASE ("InputSection::assignAnyEnum", "[estimators]") | |
| TEST_CASE ("InputSection::readXML", "[estimators]") | |
| TEST_CASE ("InputSection::InvalidElement", "[estimators]") | |
| TEST_CASE ("InputSection::init", "[estimators]") | |
| TEST_CASE ("InputSection::get", "[estimators]") | |
| TEST_CASE ("InputSection::custom", "[estimators]") | |
| std::any | makeAnotherInput (xmlNodePtr cur, std::string &value_label) |
| TEST_CASE ("InputSection::Delegate", "[estimators]") | |
| TEST_CASE ("LocalEnergyOnly", "[estimators]") | |
| TEST_CASE ("LocalEnergy", "[estimators]") | |
| TEST_CASE ("LocalEnergy with hdf5", "[estimators]") | |
| TEST_CASE ("MagnetizationDensityInput::from_xml", "[estimators]") | |
| TEST_CASE ("EstimatorManagerBase", "[estimators]") | |
| TEST_CASE ("Estimator adhoc addVector operator", "[estimators]") | |
| testing::EstimatorManagerNewTest | embt (ham, c, num_ranks) |
| embt | fakeMainScalarSamples () |
| embt | testMakeBlockAverages () |
| if (c->rank()==0) | |
| embt | fakeSomeOperatorEstimatorSamples (c->rank()) |
| embt | testReduceOperatorEstimators () |
| TEST_CASE ("MomentumDistribution::MomentumDistribution", "[estimators]") | |
| TEST_CASE ("MomentumDistribution::accumulate", "[estimators]") | |
| TEST_CASE ("MomentumDistribution::spawnCrowdClone", "[estimators]") | |
| TEST_CASE ("SpaceGrid::Construction", "[estimators]") | |
| TEST_CASE ("SpaceGrid::CYLINDRICAL", "[estimators]") | |
| TEST_CASE ("SpaceGrid::SPHERICAL", "[estimators]") | |
| TEST_CASE ("SpaceGrid::Basic", "[estimators]") | |
| TEST_CASE ("SpaceGrid::Accumulate::outside", "[estimators]") | |
| TEST_CASE ("SpaceGrid::BadPeriodic", "[estimators]") | |
| TEST_CASE ("SpaceGrid::hdf5", "[estimators]") | |
| TEST_CASE ("OneBodyDensityMatrices::OneBodyDensityMatrices", "[estimators]") | |
| TEST_CASE ("OneBodyDensityMatrices::generateSamples", "[estimators]") | |
| OneBodyDensityMatrices | original (std::move(obdmi), pset_target.getLattice(), species_set, spomap, pset_target) |
| REQUIRE (clone !=nullptr) | |
| REQUIRE (clone.get() !=&original) | |
| REQUIRE (dynamic_cast< decltype(&original)>(clone.get()) !=nullptr) | |
| TEST_CASE ("OneBodyDensityMatrices::accumulate", "[estimators]") | |
| TEST_CASE ("OneBodyDensityMatrices::evaluateMatrix", "[estimators]") | |
| TEST_CASE ("OneBodyDensityMatrices::registerAndWrite", "[estimators]") | |
| TEST_CASE ("OneBodyDensityMatricesInput::from_xml", "[estimators]") | |
| TEST_CASE ("OneBodyDensityMatricesInput::copy_construction", "[estimators]") | |
| template<typename T > | |
| std::ostream & | operator<< (std::ostream &out, const AxisGrid< T > &rhs) |
| TEMPLATE_TEST_CASE ("ParseGridInput::Good", "[estimators]", float, double) | |
| TEMPLATE_TEST_CASE ("ParseGridInput::Bad", "[estimators]", float, double) | |
| TEMPLATE_TEST_CASE ("ParseGridInput_constructors", "[estimators]", float, double) | |
| TEST_CASE ("PerParticleHamiltonianLogger_sum", "[estimators]") | |
| std::ostream & | operator<< (std::ostream &out, const testing::TestableNEReferencePoints &rhs) |
| template<typename T1 , typename T2 , unsigned D> | |
| bool | approxEquality (const TinyVector< T1, D > &val_a, const TinyVector< T2, D > &val_b) |
| ReferencePointsInput | makeTestRPI () |
| PSetsAndRefList | makePsets () |
| auto | expectedReferencePoints () |
| TEST_CASE ("ReferencePoints::DefaultConstruction", "[estimators]") | |
| TEST_CASE ("ReferencePoints::Construction", "[estimators]") | |
| TEST_CASE ("ReferencePoints::Description", "[estimators]") | |
| TEST_CASE ("ReferencePoints::HDF5", "[estimators]") | |
| TEST_CASE ("ReferencePointsInput::parseXML::valid", "[estimators]") | |
| TEST_CASE ("ReferencePointsInput::parseXML::invalid", "[estimators]") | |
| TEST_CASE ("ReferencePointsInput::makeReferencePointsInput", "[estimators]") | |
| TEST_CASE ("Scalar Estimator Input", "[estimators]") | |
| TEST_CASE ("SizeLimitedDataQueue", "[estimators]") | |
| TEST_CASE ("SpaceGridInputs::parseXML::valid", "[estimators]") | |
| TEST_CASE ("SpaceGridInputs::parseXML::invalid", "[estimators]") | |
| template<typename T > | |
| bool | checkVec (T &vec, T expected_vec) |
| TEST_CASE ("SpaceGridInputs::parseXML::axes", "[estimators]") | |
| TEST_CASE ("SpinDensityInput::readXML", "[estimators]") | |
| void | accumulateFromPsets (int ncrowds, SpinDensityNew &sdn, UPtrVector< OperatorEstBase > &crowd_sdns) |
| void | randomUpdateAccumulate (testing::RandomForTest< QMCT::RealType > &rft, UPtrVector< OperatorEstBase > &crowd_sdns) |
| REQUIRE (okay) | |
| species_set (iattribute, ispecies) | |
| SpinDensityNew (std::move(sdi), species_set) | |
| SpinDensityInput | sdi (node) |
| sdnt | testCopyConstructor (sdn) |
| TEST_CASE ("SpinDensityNew::accumulate", "[estimators]") | |
| TEST_CASE ("TraceManager", "[estimators]") | |
| TEST_CASE ("TraceManager check_trace_build", "[estimators]") | |
| template<typename T > | |
| bool | TraceSample_comp (TraceSample< T > *left, TraceSample< T > *right) |
| template void | DMCBatched::advanceWalkers< CoordsType::POS > (const StateForThread &sft, Crowd &crowd, DriverTimers &timers, DMCTimers &dmc_timers, ContextForSteps &step_context, bool recompute, bool accumulate_this_step) |
| template void | DMCBatched::advanceWalkers< CoordsType::POS_SPIN > (const StateForThread &sft, Crowd &crowd, DriverTimers &timers, DMCTimers &dmc_timers, ContextForSteps &step_context, bool recompute, bool accumulate_this_step) |
| std::ostream & | operator<< (std::ostream &o_stream, const DMCDriverInput &dmci) |
| WalkerControlBase * | createWalkerController (int nwtot, Communicate *comm, xmlNodePtr cur, bool reconfig=false) |
| function to create WalkerControlBase or its derived class More... | |
| template<class T , class TG , unsigned D> | |
| T | getDriftScale (T tau, const ParticleAttrib< TinyVector< TG, D >> &ga) |
| template<class Tt , class TG , class T , unsigned D> | |
| void | getScaledDrift (Tt tau, const TinyVector< TG, D > &qf, TinyVector< T, D > &drift) |
| evaluate a drift with a real force More... | |
| template<class Tt , class TG , class T , unsigned D> | |
| void | getScaledDriftL2 (Tt tau, const TinyVector< TG, D > &qf, const Tensor< T, D > &Dmat, TinyVector< T, D > &Kvec, TinyVector< T, D > &drift) |
| evaluate a drift with a real force with rescaling for an L2 potential More... | |
| template<class Tt , class TG , class T , unsigned D> | |
| void | getUnscaledDrift (Tt tau, const TinyVector< TG, D > &qf, TinyVector< T, D > &drift) |
| evaluate a drift with a real force and no rescaling More... | |
| template<class T , class T1 , unsigned D> | |
| T | setScaledDriftPbyPandNodeCorr (T tau, const ParticleAttrib< TinyVector< T1, D >> &qf, ParticleAttrib< TinyVector< T, D >> &drift) |
| scale drift More... | |
| template<class T , class T1 , unsigned D> | |
| T | setScaledDriftPbyPandNodeCorr (T tau_au, const std::vector< T > &massinv, const ParticleAttrib< TinyVector< T1, D >> &qf, ParticleAttrib< TinyVector< T, D >> &drift) |
| scale drift More... | |
| template<class T , class TG , unsigned D> | |
| void | setScaledDrift (T tau, const ParticleAttrib< TinyVector< TG, D >> &qf, ParticleAttrib< TinyVector< T, D >> &drift) |
| da = scaled(tau)*ga More... | |
| template<class T , unsigned D> | |
| void | setScaledDrift (T tau, ParticleAttrib< TinyVector< T, D >> &drift) |
| da = scaled(tau)*ga More... | |
| template<class T , class TG , unsigned D> | |
| void | setScaledDrift (T tau, const ParticleAttrib< TinyVector< std::complex< TG >, D >> &qf, ParticleAttrib< TinyVector< T, D >> &drift) |
| da = scaled(tau)*ga More... | |
| template<class T , class TG , unsigned D> | |
| void | assignDrift (T s, const ParticleAttrib< TinyVector< TG, D >> &ga, ParticleAttrib< TinyVector< T, D >> &da) |
| template<class T , class TG , unsigned D> | |
| void | assignDrift (T s, const ParticleAttrib< TinyVector< std::complex< TG >, D >> &ga, ParticleAttrib< TinyVector< T, D >> &da) |
| template<class T , class T1 , unsigned D> | |
| void | assignDrift (T tau_au, const std::vector< T > &massinv, const ParticleAttrib< TinyVector< T1, D >> &qf, ParticleAttrib< TinyVector< T, D >> &drift) |
| constexpr bool | operator & (DriverDebugChecks x, DriverDebugChecks y) |
| constexpr DriverDebugChecks | operator| (DriverDebugChecks x, DriverDebugChecks y) |
| constexpr DriverDebugChecks | operator~ (DriverDebugChecks x) |
| DriverDebugChecks & | operator|= (DriverDebugChecks &x, DriverDebugChecks y) |
| DriftModifierBase * | createDriftModifier (xmlNodePtr cur, const Communicate *myComm) |
| create DriftModifier More... | |
| DriftModifierBase * | createDriftModifier (const std::string &drift_modifier_str, QMCTraits::RealType unr_a) |
| std::ostream & | operator<< (std::ostream &o_stream, const QMCDriverNew &qmcd) |
| std::ostream & | operator<< (std::ostream &os, SFNBranch::VParamType &rhs) |
| std::ostream & | operator<< (std::ostream &os, SimpleFixedNodeBranch::VParamType &rhs) |
| TEST_CASE ("QMCUpdate", "[drivers]") | |
| TEST_CASE ("CloneManager", "[drivers]") | |
| TEST_CASE ("Crowd integration", "[drivers]") | |
| TEST_CASE ("Crowd redistribute walkers") | |
| TEST_CASE ("DescentEngine RMSprop update", "[drivers][descent]") | |
| This provides a basic test of the descent engine's parameter update algorithm. More... | |
| TEST_CASE ("DMC Particle-by-Particle advanceWalkers ConstantOrbital", "[drivers][dmc]") | |
| TEST_CASE ("DMC Particle-by-Particle advanceWalkers LinearOrbital", "[drivers][dmc]") | |
| TEST_CASE ("DMC", "[drivers][dmc]") | |
| TEST_CASE ("SODMC", "[drivers][dmc]") | |
| TEST_CASE ("drift pbyp and node correction real", "[drivers][drift]") | |
| TEST_CASE ("get scaled drift real", "[drivers][drift]") | |
| TEST_CASE ("solveGeneralizedEigenvalues", "[drivers]") | |
| TEST_CASE ("solveGeneralizedEigenvaluesInv", "[drivers]") | |
| TEST_CASE ("solveGeneralizedEigenvaluesCompare", "[drivers]") | |
| TEST_CASE ("EngineHandle construction", "[drivers]") | |
| This provides a basic test of constructing an EngineHandle object and checking information in it. More... | |
| TEST_CASE ("Fixed node branch", "[drivers][walker_control]") | |
| TEST_CASE ("Hybrid Engine query store","[drivers][hybrid]") | |
| This provides a basic test of the hybrid engine's check on whether vectors need to be stored. More... | |
| TEST_CASE ("selectEigenvalues", "[drivers]") | |
| TEST_CASE ("MCPopulation::createWalkers", "[particle][population]") | |
| TEST_CASE ("MCPopulation::createWalkers_walker_ids", "[particle][population]") | |
| TEST_CASE ("MCPopulation::redistributeWalkers", "[particle][population]") | |
| TEST_CASE ("updateXmlNodes", "[drivers]") | |
| TEST_CASE ("updateXmlNodes with existing element", "[drivers]") | |
| void | compute_batch_parameters (int sample_size, int batch_size, int &num_batches, int &final_batch_size) |
| Compute number of batches and final batch size given the number of samples and a batch size. More... | |
| TEST_CASE ("compute_batch_parameters", "[drivers]") | |
| TEST_CASE ("fillOverlapAndHamiltonianMatrices", "[drivers]") | |
| void | fill_from_text (int num_opt_crowds, FillData &fd) |
| TEST_CASE ("fillfromText", "[drivers]") | |
| TEST_CASE ("QMCDriverFactory create VMC Driver", "[qmcapp]") | |
| TEST_CASE ("QMCDriverFactory create VMCBatched driver", "[qmcapp]") | |
| TEST_CASE ("QMCDriverFactory create DMC driver", "[qmcapp]") | |
| TEST_CASE ("QMCDriverFactory create DMCBatched driver", "[qmcapp]") | |
| TEST_CASE ("QMCDriverInput Instantiation", "[drivers]") | |
| TEST_CASE ("QMCDriverInput readXML", "[drivers]") | |
| TEST_CASE ("QMCDriverNew tiny case", "[drivers]") | |
| TEST_CASE ("QMCDriverNew test driver operations", "[drivers]") | |
| SetupSFNBranch | setup_sfnb (pools.comm) |
| TEST_CASE ("Taus", "[Particle]") | |
| TEST_CASE ("VMC Particle-by-Particle advanceWalkers", "[drivers][vmc]") | |
| TEST_CASE ("VMC", "[drivers][vmc]") | |
| TEST_CASE ("SOVMC", "[drivers][vmc]") | |
| TEST_CASE ("SOVMC-alle", "[drivers][vmc]") | |
| TEST_CASE ("VMCBatched::calc_default_local_walkers", "[drivers]") | |
| TEST_CASE ("VMCDriverInput readXML", "[drivers]") | |
| TEST_CASE ("VMCFactory Instantiation", "[drivers]") | |
| void | output_vector (const std::string &name, std::vector< int > &vec) |
| TEST_CASE ("Walker control assign walkers", "[drivers][walker_control]") | |
| TEST_CASE ("WalkerControl round trip index conversions", "[drivers][walker_control]") | |
| TEST_CASE ("Walker control assign walkers odd ranks", "[drivers][walker_control]") | |
| TEST_CASE ("WalkerControl::determineNewWalkerPopulation", "[drivers][walker_control]") | |
| TEST_CASE ("WalkerLogCollector::collect", "[estimators]") | |
| TEST_CASE ("WFOptDriverInput Instantiation", "[drivers]") | |
| TEST_CASE ("WFOptDriverInput readXML", "[drivers]") | |
| template void | VMCBatched::advanceWalkers< CoordsType::POS > (const StateForThread &sft, Crowd &crowd, QMCDriverNew::DriverTimers &timers, ContextForSteps &step_context, bool recompute, bool accumulate_this_step) |
| template void | VMCBatched::advanceWalkers< CoordsType::POS_SPIN > (const StateForThread &sft, Crowd &crowd, QMCDriverNew::DriverTimers &timers, ContextForSteps &step_context, bool recompute, bool accumulate_this_step) |
| std::ostream & | operator<< (std::ostream &o_stream, const VMCBatched &vmc_batched) |
| std::ostream & | operator<< (std::ostream &o_stream, const VMCDriverInput &vmci) |
| template<typename T , unsigned D> | |
| void | randomize (ParticleAttrib< TinyVector< T, D >> &displ, T fac) |
| std::unique_ptr< QMCFixedSampleLinearOptimizeBatched > | QMCWFOptLinearFactoryNew (xmlNodePtr cur, const ProjectData &project_data, const std::optional< EstimatorManagerInput > &global_emi, WalkerConfigurations &wc, MCPopulation &&pop, SampleStack &samples, Communicate *comm) |
| void | get_gridinfo_from_posgrid (const std::vector< PosType > &posgridlist, const IndexType &axis, RealType &lx, RealType &rx, RealType &dx, IndexType &Nx) |
| void | getStats (const std::vector< RealType > &vals, RealType &avg, RealType &err, int start=0) |
| Simpleaverage and error estimate. More... | |
| int | estimateEquilibration (const std::vector< RealType > &vals, RealType frac=0.75) |
| estimate equilibration of block data More... | |
| TEST_CASE ("FS parse Sk file", "[tools]") | |
| TEST_CASE ("FS evaluate", "[tools]") | |
| std::string | lowerCase (const std::string_view s) |
| ++17 More... | |
| template<typename T > | |
| T | string2Real (const std::string_view svalue) |
| alternate to string2real calls c++ string to real conversion based on T's precision template<typename T> More... | |
| template<typename T > | |
| T | string2Int (const std::string_view svalue) |
| alternate to string2real calls c++ string to real conversion based on T's precision template<typename T> More... | |
Variables | |
| hdf_error_suppression | hide_hdf_errors |
| Suppress HDF5 warning and error messages. More... | |
| bool | timer_max_level_exceeded = false |
| QMCState | qmc_common |
| a unique QMCState during a run More... | |
| RNGThreadSafe< FakeRandom< OHMMS_PRECISION_FULL > > | fake_random_global |
| RNGThreadSafe< RandomGenerator > | random_global |
| RunTimeManager< ChronoClock > | run_time_manager |
| static std::ostringstream | summary_out |
| static std::ostringstream | app_out |
| static std::ostringstream | err_out |
| static std::ostringstream | debug_out |
| TimerNameList_t< TestTimer > | TestTimerNames = {{MyTimer1, "Timer name 1"}, {MyTimer2, "Timer name 2"}} |
| std::atomic< size_t > | CUDAallocator_device_mem_allocated |
| std::atomic< size_t > | dual_device_mem_allocated |
| std::atomic< size_t > | OMPallocator_device_mem_allocated |
| std::atomic< size_t > | SYCLallocator_device_mem_allocated |
| auto | checkRs |
| MCCoords< CoordsType::POS > | mc_coords_rs (size_test) |
| MCCoords< CoordsType::POS_SPIN > | mc_coords_rsspins (size_test) |
| int | offset_for_rs = (3 * size_test) + (3 * size_test) % 2 |
| constexpr RealType | no_energy_tol = std::numeric_limits<RealType>::max() |
| constexpr int | no_index = -1 |
| const int | inone = std::numeric_limits<int>::min() |
| const RealType | rnone = std::numeric_limits<RealType>::max() |
| const std::string & | snone = "none" |
| int | n = 4 |
| int | lda = 4 |
| auto & | hstream = cuda_handles->hstream |
| int | batch_size = 2 |
| std::vector< StdComp, CUDAHostAllocator< StdComp > > | lu |
| std::vector< StdComp, CUDAHostAllocator< StdComp > > | lu2 |
| lus [0] = dev_lu.data() | |
| std::vector< int, CUDAHostAllocator< int > > | pivots = {3, 4, 3, 4, 3, 4, 4, 4} |
| std::vector< double, CUDAHostAllocator< double > > | M_vec {2, 5, 7, 5, 5, 2, 5, 4, 8, 2, 6, 4, 7, 8, 6, 8} |
| std::vector< double, CUDAHostAllocator< double > > | M2_vec {6, 5, 7, 5, 2, 2, 5, 4, 8, 2, 6, 4, 3, 8, 6, 8} |
| std::vector< double *, CUDAHostAllocator< double * > > | Ms {devM_vec.data(), devM2_vec.data()} |
| std::vector< int > | real_pivot {3, 3, 4, 4, 3, 3, 3, 4} |
| auto | checkArray |
| auto | check_matrix_result = checkMatrix(lu_mat, M_mat) |
| const std::string | identity_coeff |
| const std::string | coeff_rot_by_point1 |
| const std::string | coeff_rot_by_point2 |
| const std::string | coeff_rot_by_point05 |
| static const std::vector< std::string > | suffixes |
| static const TimerNameList_t< DMTimers > | DMTimerNames |
| hdf_archive | hFile |
| ObservableHelper | oh {hdf_path{"u"}} |
| std::vector< int > | dims = {10, 10} |
| float | propertyFloat = 10.f |
| Tensor< float, OHMMS_DIM > | propertyTensor |
| Matrix< float > | propertyMatrix |
| TinyVector< float, OHMMS_DIM > | propertyTinyVector |
| std::vector< float > | propertyVector |
| std::vector< TinyVector< float, OHMMS_DIM > > | propertyVectorTinyVector |
| constexpr bool | generate_test_data = false |
| set to true to generate new random R for particles. More... | |
| QMCHamiltonian | ham = *(hamiltonian_pool.getPrimary()) |
| testing::EstimatorManagerNewTest | embt (ham, c, 1) |
| Communicate * | comm = OHMMS::Controller |
| Libxml2Document | estimators_doc = createEstimatorManagerNewVMCInputXML() |
| auto | particle_pool = MinimalParticlePool::make_diamondC_1x1x1(comm) |
| auto | wavefunction_pool |
| auto & | pset = *(particle_pool.getParticleSet("e")) |
| auto | hamiltonian_pool = MinimalHamiltonianPool::make_hamWithEE(comm, particle_pool, wavefunction_pool) |
| auto & | twf = *(wavefunction_pool.getWaveFunction("wavefunction")) |
| EstimatorManagerNew | emn (comm, std::move(emi), ham, pset, twf) |
| EstimatorManagerNewTestAccess | emnta (emn) |
| Libxml2Document | estimators_doc2 = createEstimatorManagerNewInputXML() |
| EstimatorManagerNew | emn2 (comm, std::move(emi2), ham, pset, twf) |
| EstimatorManagerNewTestAccess | emnta2 (emn2) |
| std::unordered_map< std::string, std::any > | lookup_input_enum_value |
| int | num_ranks = c->size() |
| std::vector< QMCTraits::RealType > | good_data = embt.generateGoodOperatorData(num_ranks) |
| constexpr bool | dump_obdm = false |
| ProjectData | test_project ("test", ProjectData::DriverVersion::BATCH) |
| auto & | pset_target = *(particle_pool.getParticleSet("e")) |
| auto & | species_set = pset_target.getSpeciesSet() |
| auto & | spomap = wavefunction_pool.getWaveFunction("wavefunction")->getSPOMap() |
| Libxml2Document | doc |
| bool | okay = doc.parseFromString(Input::xml[Input::valid::VANILLA]) |
| if(!okay) throw std xmlNodePtr | node = doc.getRoot() |
| OneBodyDensityMatricesInput | obdmi (node) |
| auto | clone = original.spawnCrowdClone() |
| SpinDensityInput | sdi (node) |
| int | ispecies = species_set.addSpecies("C") |
| int | iattribute = species_set.addAttribute("membersize") |
| SpinDensityInput | sdi_copy = sdi |
| CrystalLattice< OHMMS_PRECISION, OHMMS_DIM > | lattice = testing::makeTestLattice() |
| SpinDensityNew | sdn (std::move(sdi), lattice, species_set) |
| OEBAccessor | oeba (sdn) = 1.0 |
| SpinDensityNewTests | sdnt |
| SpinDensityNew | original (std::move(sdi), lattice, species_set) |
| const unsigned int | DMAX = 4 |
| TimerNameList_t< DMCTimers > | DMCTimerNames |
| const TimerNameList_t< SODMCTimers > | SODMCTimerNames |
| TimerNameList_t< WC_Timers > | WalkerControlTimerNames |
| TimerNameList_t< DMC_MPI_Timers > | DMCMPITimerNames |
| const std::map< std::string, OptimizerType > | OptimizerNames |
| SetupPools | pools |
| std::unique_ptr< SFNBranch > | sfnb |
| SetupSimpleFixedNodeBranch | setup_sfnb |
Convenience templated type aliases | |
when feasible do not use previous aliases in following that way one line can be scanned to understand a single alias see: UPtrVector | |
| template<typename T > | |
| using | RefVector = std::vector< std::reference_wrapper< T > > |
| template<typename T > | |
| using | UPtr = std::unique_ptr< T > |
| template<typename T > | |
| using | UPtrVector = std::vector< std::unique_ptr< T > > |
| template<class TR , class T > | |
| static RefVector< TR > | makeRefVector (std::vector< T > &vec_list) |
| }@ More... | |
| template<class T > | |
| static RefVector< T > | convertUPtrToRefVector (const UPtrVector< T > &ptr_list) |
| convert a vector of std::unique_ptrs<T> to a refvector<T> More... | |
| template<class T2 , class T > | |
| static RefVector< T2 > | convertUPtrToRefVector (const UPtrVector< T > &ptr_list) |
| convert a vector of std::unique_ptrs<T> to a refvector<T2> the static assert prevents ambiguity between this function and the above when T is a derived type of T2. More... | |
| template<class T > | |
| static RefVector< T > | convertPtrToRefVector (const std::vector< T *> &ptr_list) |
| template<class T > | |
| static std::vector< T * > | convert_ref_to_ptr_list (const std::vector< std::reference_wrapper< T >> &ref_list) |
| template<class T > | |
| static std::vector< T * > | convertUPtrToPtrVector (const UPtrVector< T > &uptr_list) |
| template<class T > | |
| static RefVector< T > | convertUPtrToRefVectorSubset (const UPtrVector< T > &ptr_list, int offset, int len) |
| template<class T > | |
| static RefVector< T > | convertPtrToRefVectorSubset (const std::vector< T *> &ptr_list, int offset, int len) |
| template<class T > | |
| static RefVector< T > | convertRefVectorToRefVectorSubset (const RefVector< T > &ref_list, int offset, int len) |
helper functions for EinsplineSetBuilder
If defined, use recursive method to build the basis set for each position.
performance improvement is questionable: load vs sin/cos
| struct qmcplusplus::AndCombine |
| struct qmcplusplus::ao_traits |
traits for a localized basis set; used by createBasisSet
T radial function value type ORBT orbital value type, can be complex ROT {0=numuerica;, 1=gto; 2=sto} SH {0=cartesian, 1=spherical} If too confusing, inroduce enumeration.
Definition at line 52 of file LCAOrbitalBuilder.cpp.
 Collaboration diagram for ao_traits< T, ORBT, ROT, SH >:
Collaboration diagram for ao_traits< T, ORBT, ROT, SH >:| struct qmcplusplus::ao_traits< T, ORBT, 0, 0 > |
specialization for numerical-cartesian AO
Definition at line 57 of file LCAOrbitalBuilder.cpp.
 Collaboration diagram for ao_traits< T, ORBT, 0, 0 >:
Collaboration diagram for ao_traits< T, ORBT, 0, 0 >:| Class Members | ||
|---|---|---|
| typedef SoaCartesianTensor< T > | angular_type | |
| typedef SoaAtomicBasisSet< radial_type, angular_type > | ao_type | |
| typedef SoaLocalizedBasisSet< ao_type, ORBT > | basis_type | |
| typedef MultiQuinticSpline1D< T > | radial_type | |
| struct qmcplusplus::ao_traits< T, ORBT, 0, 1 > |
specialization for numerical-spherical AO
Definition at line 67 of file LCAOrbitalBuilder.cpp.
 Collaboration diagram for ao_traits< T, ORBT, 0, 1 >:
Collaboration diagram for ao_traits< T, ORBT, 0, 1 >:| Class Members | ||
|---|---|---|
| typedef SoaSphericalTensor< T > | angular_type | |
| typedef SoaAtomicBasisSet< radial_type, angular_type > | ao_type | |
| typedef SoaLocalizedBasisSet< ao_type, ORBT > | basis_type | |
| typedef MultiQuinticSpline1D< T > | radial_type | |
| struct qmcplusplus::ao_traits< T, ORBT, 1, 0 > |
specialization for GTO-cartesian AO
Definition at line 77 of file LCAOrbitalBuilder.cpp.
 Collaboration diagram for ao_traits< T, ORBT, 1, 0 >:
Collaboration diagram for ao_traits< T, ORBT, 1, 0 >:| Class Members | ||
|---|---|---|
| typedef SoaCartesianTensor< T > | angular_type | |
| typedef SoaAtomicBasisSet< radial_type, angular_type > | ao_type | |
| typedef SoaLocalizedBasisSet< ao_type, ORBT > | basis_type | |
| typedef MultiFunctorAdapter< GaussianCombo< T > > | radial_type | |
| struct qmcplusplus::ao_traits< T, ORBT, 1, 1 > |
specialization for GTO-cartesian AO
Definition at line 87 of file LCAOrbitalBuilder.cpp.
 Collaboration diagram for ao_traits< T, ORBT, 1, 1 >:
Collaboration diagram for ao_traits< T, ORBT, 1, 1 >:| Class Members | ||
|---|---|---|
| typedef SoaSphericalTensor< T > | angular_type | |
| typedef SoaAtomicBasisSet< radial_type, angular_type > | ao_type | |
| typedef SoaLocalizedBasisSet< ao_type, ORBT > | basis_type | |
| typedef MultiFunctorAdapter< GaussianCombo< T > > | radial_type | |
| struct qmcplusplus::ao_traits< T, ORBT, 2, 0 > |
specialization for STO-cartesian AO
Definition at line 107 of file LCAOrbitalBuilder.cpp.
 Collaboration diagram for ao_traits< T, ORBT, 2, 0 >:
Collaboration diagram for ao_traits< T, ORBT, 2, 0 >:| Class Members | ||
|---|---|---|
| typedef SoaCartesianTensor< T > | angular_type | |
| typedef SoaAtomicBasisSet< radial_type, angular_type > | ao_type | |
| typedef SoaLocalizedBasisSet< ao_type, ORBT > | basis_type | |
| typedef MultiFunctorAdapter< SlaterCombo< T > > | radial_type | |
| struct qmcplusplus::ao_traits< T, ORBT, 2, 1 > |
specialization for STO-spherical AO
Definition at line 97 of file LCAOrbitalBuilder.cpp.
 Collaboration diagram for ao_traits< T, ORBT, 2, 1 >:
Collaboration diagram for ao_traits< T, ORBT, 2, 1 >:| Class Members | ||
|---|---|---|
| typedef SoaSphericalTensor< T > | angular_type | |
| typedef SoaAtomicBasisSet< radial_type, angular_type > | ao_type | |
| typedef SoaLocalizedBasisSet< ao_type, ORBT > | basis_type | |
| typedef MultiFunctorAdapter< SlaterCombo< T > > | radial_type | |
| struct qmcplusplus::ApplyBConds |
Dummy template class to apply boundary conditions.
Definition at line 181 of file FastParticleOperators.h.
 Collaboration diagram for ApplyBConds< T1, T2, D >:
Collaboration diagram for ApplyBConds< T1, T2, D >:| struct qmcplusplus::astring |
Definition at line 172 of file string_utils.h.
 Collaboration diagram for astring:
Collaboration diagram for astring:| Class Members | ||
|---|---|---|
| string | s | |
| struct qmcplusplus::BinaryReturn |
Definition at line 430 of file TypeComputations.h.
 Collaboration diagram for BinaryReturn< T1, T2, Op >:
Collaboration diagram for BinaryReturn< T1, T2, Op >:| Class Members | ||
|---|---|---|
| typedef typename Promote< T1, T2 >::Type_t | Type_t | |
| struct qmcplusplus::BinaryReturn< std::complex< T1 >, T1, OpMultiply > |
Definition at line 26 of file TinyVectorOps.h.
 Collaboration diagram for BinaryReturn< std::complex< T1 >, T1, OpMultiply >:
Collaboration diagram for BinaryReturn< std::complex< T1 >, T1, OpMultiply >:| Class Members | ||
|---|---|---|
| typedef complex< T1 > | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, std::complex< T1 >, OpMultiply > |
Definition at line 20 of file TinyVectorOps.h.
 Collaboration diagram for BinaryReturn< T1, std::complex< T1 >, OpMultiply >:
Collaboration diagram for BinaryReturn< T1, std::complex< T1 >, OpMultiply >:| Class Members | ||
|---|---|---|
| typedef complex< T1 > | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpAddAssign > |
Definition at line 575 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpAddAssign >:
Collaboration diagram for BinaryReturn< T1, T2, OpAddAssign >:| Class Members | ||
|---|---|---|
| typedef T1 & | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpAnd > |
Definition at line 510 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpAnd >:
Collaboration diagram for BinaryReturn< T1, T2, OpAnd >:| Class Members | ||
|---|---|---|
| typedef bool | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpAssign > |
Definition at line 744 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpAssign >:
Collaboration diagram for BinaryReturn< T1, T2, OpAssign >:| Class Members | ||
|---|---|---|
| typedef T1 & | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpBitwiseAndAssign > |
Definition at line 677 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpBitwiseAndAssign >:
Collaboration diagram for BinaryReturn< T1, T2, OpBitwiseAndAssign >:| Class Members | ||
|---|---|---|
| typedef T1 & | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpBitwiseOrAssign > |
Definition at line 660 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpBitwiseOrAssign >:
Collaboration diagram for BinaryReturn< T1, T2, OpBitwiseOrAssign >:| Class Members | ||
|---|---|---|
| typedef T1 & | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpBitwiseXorAssign > |
Definition at line 694 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpBitwiseXorAssign >:
Collaboration diagram for BinaryReturn< T1, T2, OpBitwiseXorAssign >:| Class Members | ||
|---|---|---|
| typedef T1 & | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpDivideAssign > |
Definition at line 626 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpDivideAssign >:
Collaboration diagram for BinaryReturn< T1, T2, OpDivideAssign >:| Class Members | ||
|---|---|---|
| typedef T1 & | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpEQ > |
Definition at line 478 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpEQ >:
Collaboration diagram for BinaryReturn< T1, T2, OpEQ >:| Class Members | ||
|---|---|---|
| typedef bool | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpGE > |
Definition at line 462 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpGE >:
Collaboration diagram for BinaryReturn< T1, T2, OpGE >:| Class Members | ||
|---|---|---|
| typedef bool | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpGT > |
Definition at line 446 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpGT >:
Collaboration diagram for BinaryReturn< T1, T2, OpGT >:| Class Members | ||
|---|---|---|
| typedef bool | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpLE > |
Definition at line 430 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpLE >:
Collaboration diagram for BinaryReturn< T1, T2, OpLE >:| Class Members | ||
|---|---|---|
| typedef bool | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpLeftShift > |
Definition at line 542 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpLeftShift >:
Collaboration diagram for BinaryReturn< T1, T2, OpLeftShift >:| Class Members | ||
|---|---|---|
| typedef T1 | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpLeftShiftAssign > |
Definition at line 711 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpLeftShiftAssign >:
Collaboration diagram for BinaryReturn< T1, T2, OpLeftShiftAssign >:| Class Members | ||
|---|---|---|
| typedef T1 & | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpLT > |
Definition at line 414 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpLT >:
Collaboration diagram for BinaryReturn< T1, T2, OpLT >:| Class Members | ||
|---|---|---|
| typedef bool | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpModAssign > |
Definition at line 643 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpModAssign >:
Collaboration diagram for BinaryReturn< T1, T2, OpModAssign >:| Class Members | ||
|---|---|---|
| typedef T1 & | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpMultiplyAssign > |
Definition at line 609 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpMultiplyAssign >:
Collaboration diagram for BinaryReturn< T1, T2, OpMultiplyAssign >:| Class Members | ||
|---|---|---|
| typedef T1 & | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpNE > |
Definition at line 494 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpNE >:
Collaboration diagram for BinaryReturn< T1, T2, OpNE >:| Class Members | ||
|---|---|---|
| typedef bool | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpOr > |
Definition at line 526 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpOr >:
Collaboration diagram for BinaryReturn< T1, T2, OpOr >:| Class Members | ||
|---|---|---|
| typedef bool | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpRightShift > |
Definition at line 558 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpRightShift >:
Collaboration diagram for BinaryReturn< T1, T2, OpRightShift >:| Class Members | ||
|---|---|---|
| typedef T1 | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpRightShiftAssign > |
Definition at line 728 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpRightShiftAssign >:
Collaboration diagram for BinaryReturn< T1, T2, OpRightShiftAssign >:| Class Members | ||
|---|---|---|
| typedef T1 & | Type_t | |
| struct qmcplusplus::BinaryReturn< T1, T2, OpSubtractAssign > |
Definition at line 592 of file OperatorTags.h.
 Collaboration diagram for BinaryReturn< T1, T2, OpSubtractAssign >:
Collaboration diagram for BinaryReturn< T1, T2, OpSubtractAssign >:| Class Members | ||
|---|---|---|
| typedef T1 & | Type_t | |
| struct qmcplusplus::CheckMatrixResult |
Definition at line 43 of file checkMatrix.hpp.
 Collaboration diagram for CheckMatrixResult:
Collaboration diagram for CheckMatrixResult:| Class Members | ||
|---|---|---|
| bool | result | |
| string | result_message | |
| struct qmcplusplus::Combine2 |
Definition at line 93 of file Combiners.h.
 Collaboration diagram for Combine2< A, B, Op, Tag >:
Collaboration diagram for Combine2< A, B, Op, Tag >:| struct qmcplusplus::const_traits |
Definition at line 32 of file determinant_operators.h.
 Collaboration diagram for const_traits< T >:
Collaboration diagram for const_traits< T >:| struct qmcplusplus::container |
Definition at line 23 of file test_deep_copy.cpp.
 Collaboration diagram for container:
Collaboration diagram for container:| Class Members | ||
|---|---|---|
| double * | data | |
| int | size | |
| struct qmcplusplus::ConvertPosUnit |
Dummy template class to be specialized.
Definition at line 31 of file FastParticleOperators.h.
 Collaboration diagram for ConvertPosUnit< T1, T2, D >:
Collaboration diagram for ConvertPosUnit< T1, T2, D >:| struct qmcplusplus::CSFData |
| class qmcplusplus::CustomizedMatrixDet |
Definition at line 151 of file SmallMatrixDetCalculator.h.
 Collaboration diagram for CustomizedMatrixDet< NEXCITED >:
Collaboration diagram for CustomizedMatrixDet< NEXCITED >:| struct qmcplusplus::DecrementLeaf |
| struct qmcplusplus::DereferenceLeaf |
| struct qmcplusplus::DetInverterSelector |
Definition at line 61 of file DiracDeterminantBatched.h.
 Collaboration diagram for DetInverterSelector< UEPL, FPVT >:
Collaboration diagram for DetInverterSelector< UEPL, FPVT >:| Class Members | ||
|---|---|---|
| typedef DiracMatrixComputeOMPTarget< FPVT > | Inverter | |
| struct qmcplusplus::double_tag |
Definition at line 37 of file test_TrialWaveFunction_diamondC_2x1x1.cpp.
 Collaboration diagram for double_tag:
Collaboration diagram for double_tag:| struct qmcplusplus::FillData |
Definition at line 18 of file FillData.h.
 Collaboration diagram for FillData:
Collaboration diagram for FillData:| Class Members | ||
|---|---|---|
| Matrix< double > | derivRecords | |
| vector< double > | energy_new | |
| Matrix< double > | ham_gold | |
| Matrix< double > | HDerivRecords | |
| int | numParam | |
| int | numSamples | |
| Matrix< double > | ovlp_gold | |
| vector< double > | reweight | |
| double | sum_e_wgt | |
| double | sum_esq_wgt | |
| double | sum_wgt | |
| struct qmcplusplus::float_tag |
Definition at line 39 of file test_TrialWaveFunction_diamondC_2x1x1.cpp.
 Collaboration diagram for float_tag:
Collaboration diagram for float_tag:| struct qmcplusplus::IncrementLeaf |
| class qmcplusplus::JastrowTypeHelper |
Definition at line 41 of file RadialJastrowBuilder.cpp.
 Collaboration diagram for JastrowTypeHelper< RadFuncType, Implementation >:
Collaboration diagram for JastrowTypeHelper< RadFuncType, Implementation >:| Class Members | ||
|---|---|---|
| typedef J1Spin< RadFuncType > | J1SpinType | |
| typedef J1OrbitalSoA< RadFuncType > | J1Type | |
| typedef TwoBodyJastrow< RadFuncType > | J2Type | |
| class qmcplusplus::JastrowTypeHelper< BsplineFunctor< RadialJastrowBuilder::RealType >, RadialJastrowBuilder::detail::OMPTARGET > |
Definition at line 50 of file RadialJastrowBuilder.cpp.
 Collaboration diagram for JastrowTypeHelper< BsplineFunctor< RadialJastrowBuilder::RealType >, RadialJastrowBuilder::detail::OMPTARGET >:
Collaboration diagram for JastrowTypeHelper< BsplineFunctor< RadialJastrowBuilder::RealType >, RadialJastrowBuilder::detail::OMPTARGET >:| Class Members | ||
|---|---|---|
| typedef TwoBodyJastrow< RadFuncType > | J2Type | |
| typedef BsplineFunctor< RealType > | RadFuncType | |
| struct qmcplusplus::LatticeAnalyzer |
generic class to analyze a Lattice
Definition at line 43 of file LatticeAnalyzer.h.
 Collaboration diagram for LatticeAnalyzer< T, D >:
Collaboration diagram for LatticeAnalyzer< T, D >:| struct qmcplusplus::LeafFunctor |
Definition at line 76 of file Functors.h.
 Collaboration diagram for LeafFunctor< LeafType, LeafTag >:
Collaboration diagram for LeafFunctor< LeafType, LeafTag >:| struct qmcplusplus::MCCoords |
Definition at line 31 of file MCCoords.hpp.
 Collaboration diagram for MCCoords< MCT >:
Collaboration diagram for MCCoords< MCT >:| struct qmcplusplus::MCDataType |
Monte Carlo Data of an ensemble.
The quantities are shared by all the nodes in a group
Definition at line 41 of file WalkerConfigurations.h.
 Inheritance diagram for MCDataType< T >:
Inheritance diagram for MCDataType< T >: Collaboration diagram for MCDataType< T >:
Collaboration diagram for MCDataType< T >:| Class Members | ||
|---|---|---|
| T | AlternateEnergy | |
| T | Energy | |
| T | LivingFraction | |
| T | NumSamples | |
| T | R2Accepted | |
| T | R2Proposed | |
| T | RNSamples | |
| T | Variance | |
| T | Weight | |
| struct qmcplusplus::NullCombine |
| struct qmcplusplus::OffloadSwitches |
Definition at line 42 of file test_TrialWaveFunction_diamondC_2x1x1.cpp.
 Collaboration diagram for OffloadSwitches:
Collaboration diagram for OffloadSwitches:| Class Members | ||
|---|---|---|
| bool | jas | |
| bool | spo | |
| struct qmcplusplus::OpCombine |
| struct qmcplusplus::OrCombine |
| struct qmcplusplus::OTAssign |
Definition at line 42 of file OhmmsTinyMeta.h.
 Collaboration diagram for OTAssign< T1, T2, OP >:
Collaboration diagram for OTAssign< T1, T2, OP >:| struct qmcplusplus::OTBinary |
Definition at line 45 of file OhmmsTinyMeta.h.
 Collaboration diagram for OTBinary< T1, T2, OP >:
Collaboration diagram for OTBinary< T1, T2, OP >:| struct qmcplusplus::OTCross |
Definition at line 402 of file TinyVectorOps.h.
 Collaboration diagram for OTCross< T1, T2 >:
Collaboration diagram for OTCross< T1, T2 >:| struct qmcplusplus::OTDot |
Definition at line 50 of file OhmmsTinyMeta.h.
 Collaboration diagram for OTDot< T1, T2 >:
Collaboration diagram for OTDot< T1, T2 >:| struct qmcplusplus::OuterProduct |
Definition at line 606 of file TinyVectorTensorOps.h.
 Collaboration diagram for OuterProduct< T1, T2 >:
Collaboration diagram for OuterProduct< T1, T2 >:| struct qmcplusplus::PAOps |
generic PAOps
Definition at line 223 of file ParticleAttribOps.h.
 Collaboration diagram for PAOps< T, D, T1 >:
Collaboration diagram for PAOps< T, D, T1 >:| struct qmcplusplus::printTinyVector |
Definition at line 220 of file TinyVector.h.
 Collaboration diagram for printTinyVector< T >:
Collaboration diagram for printTinyVector< T >:| struct qmcplusplus::Promote |
Definition at line 83 of file TypeComputations.h.
 Collaboration diagram for Promote< T1, T2 >:
Collaboration diagram for Promote< T1, T2 >:| Class Members | ||
|---|---|---|
| typedef T1 | Type_t | |
| struct qmcplusplus::Promote< bool, bool > |
Definition at line 91 of file TypeComputations.h.
 Collaboration diagram for Promote< bool, bool >:
Collaboration diagram for Promote< bool, bool >:| Class Members | ||
|---|---|---|
| typedef bool | Type_t | |
| struct qmcplusplus::Promote< bool, char > |
Definition at line 97 of file TypeComputations.h.
 Collaboration diagram for Promote< bool, char >:
Collaboration diagram for Promote< bool, char >:| Class Members | ||
|---|---|---|
| typedef char | Type_t | |
| struct qmcplusplus::Promote< bool, double > |
Definition at line 127 of file TypeComputations.h.
 Collaboration diagram for Promote< bool, double >:
Collaboration diagram for Promote< bool, double >:| Class Members | ||
|---|---|---|
| typedef double | Type_t | |
| struct qmcplusplus::Promote< bool, float > |
Definition at line 121 of file TypeComputations.h.
 Collaboration diagram for Promote< bool, float >:
Collaboration diagram for Promote< bool, float >:| Class Members | ||
|---|---|---|
| typedef float | Type_t | |
| struct qmcplusplus::Promote< bool, int > |
Definition at line 109 of file TypeComputations.h.
 Collaboration diagram for Promote< bool, int >:
Collaboration diagram for Promote< bool, int >:| Class Members | ||
|---|---|---|
| typedef int | Type_t | |
| struct qmcplusplus::Promote< bool, long > |
Definition at line 115 of file TypeComputations.h.
 Collaboration diagram for Promote< bool, long >:
Collaboration diagram for Promote< bool, long >:| Class Members | ||
|---|---|---|
| typedef long | Type_t | |
| struct qmcplusplus::Promote< bool, short > |
Definition at line 103 of file TypeComputations.h.
 Collaboration diagram for Promote< bool, short >:
Collaboration diagram for Promote< bool, short >:| Class Members | ||
|---|---|---|
| typedef short | Type_t | |
| struct qmcplusplus::Promote< char, bool > |
Definition at line 135 of file TypeComputations.h.
 Collaboration diagram for Promote< char, bool >:
Collaboration diagram for Promote< char, bool >:| Class Members | ||
|---|---|---|
| typedef char | Type_t | |
| struct qmcplusplus::Promote< char, char > |
Definition at line 141 of file TypeComputations.h.
 Collaboration diagram for Promote< char, char >:
Collaboration diagram for Promote< char, char >:| Class Members | ||
|---|---|---|
| typedef char | Type_t | |
| struct qmcplusplus::Promote< char, double > |
Definition at line 171 of file TypeComputations.h.
 Collaboration diagram for Promote< char, double >:
Collaboration diagram for Promote< char, double >:| Class Members | ||
|---|---|---|
| typedef double | Type_t | |
| struct qmcplusplus::Promote< char, float > |
Definition at line 165 of file TypeComputations.h.
 Collaboration diagram for Promote< char, float >:
Collaboration diagram for Promote< char, float >:| Class Members | ||
|---|---|---|
| typedef float | Type_t | |
| struct qmcplusplus::Promote< char, int > |
Definition at line 153 of file TypeComputations.h.
 Collaboration diagram for Promote< char, int >:
Collaboration diagram for Promote< char, int >:| Class Members | ||
|---|---|---|
| typedef int | Type_t | |
| struct qmcplusplus::Promote< char, long > |
Definition at line 159 of file TypeComputations.h.
 Collaboration diagram for Promote< char, long >:
Collaboration diagram for Promote< char, long >:| Class Members | ||
|---|---|---|
| typedef long | Type_t | |
| struct qmcplusplus::Promote< char, short > |
Definition at line 147 of file TypeComputations.h.
 Collaboration diagram for Promote< char, short >:
Collaboration diagram for Promote< char, short >:| Class Members | ||
|---|---|---|
| typedef short | Type_t | |
| struct qmcplusplus::Promote< double, bool > |
Definition at line 355 of file TypeComputations.h.
 Collaboration diagram for Promote< double, bool >:
Collaboration diagram for Promote< double, bool >:| Class Members | ||
|---|---|---|
| typedef double | Type_t | |
| struct qmcplusplus::Promote< double, char > |
Definition at line 361 of file TypeComputations.h.
 Collaboration diagram for Promote< double, char >:
Collaboration diagram for Promote< double, char >:| Class Members | ||
|---|---|---|
| typedef double | Type_t | |
| struct qmcplusplus::Promote< double, double > |
Definition at line 391 of file TypeComputations.h.
 Collaboration diagram for Promote< double, double >:
Collaboration diagram for Promote< double, double >:| Class Members | ||
|---|---|---|
| typedef double | Type_t | |
| struct qmcplusplus::Promote< double, float > |
Definition at line 385 of file TypeComputations.h.
 Collaboration diagram for Promote< double, float >:
Collaboration diagram for Promote< double, float >:| Class Members | ||
|---|---|---|
| typedef double | Type_t | |
| struct qmcplusplus::Promote< double, int > |
Definition at line 373 of file TypeComputations.h.
 Collaboration diagram for Promote< double, int >:
Collaboration diagram for Promote< double, int >:| Class Members | ||
|---|---|---|
| typedef double | Type_t | |
| struct qmcplusplus::Promote< double, long > |
Definition at line 379 of file TypeComputations.h.
 Collaboration diagram for Promote< double, long >:
Collaboration diagram for Promote< double, long >:| Class Members | ||
|---|---|---|
| typedef double | Type_t | |
| struct qmcplusplus::Promote< double, short > |
Definition at line 367 of file TypeComputations.h.
 Collaboration diagram for Promote< double, short >:
Collaboration diagram for Promote< double, short >:| Class Members | ||
|---|---|---|
| typedef double | Type_t | |
| struct qmcplusplus::Promote< float, bool > |
Definition at line 311 of file TypeComputations.h.
 Collaboration diagram for Promote< float, bool >:
Collaboration diagram for Promote< float, bool >:| Class Members | ||
|---|---|---|
| typedef float | Type_t | |
| struct qmcplusplus::Promote< float, char > |
Definition at line 317 of file TypeComputations.h.
 Collaboration diagram for Promote< float, char >:
Collaboration diagram for Promote< float, char >:| Class Members | ||
|---|---|---|
| typedef float | Type_t | |
| struct qmcplusplus::Promote< float, double > |
Definition at line 347 of file TypeComputations.h.
 Collaboration diagram for Promote< float, double >:
Collaboration diagram for Promote< float, double >:| Class Members | ||
|---|---|---|
| typedef double | Type_t | |
| struct qmcplusplus::Promote< float, float > |
Definition at line 341 of file TypeComputations.h.
 Collaboration diagram for Promote< float, float >:
Collaboration diagram for Promote< float, float >:| Class Members | ||
|---|---|---|
| typedef float | Type_t | |
| struct qmcplusplus::Promote< float, int > |
Definition at line 329 of file TypeComputations.h.
 Collaboration diagram for Promote< float, int >:
Collaboration diagram for Promote< float, int >:| Class Members | ||
|---|---|---|
| typedef float | Type_t | |
| struct qmcplusplus::Promote< float, long > |
Definition at line 335 of file TypeComputations.h.
 Collaboration diagram for Promote< float, long >:
Collaboration diagram for Promote< float, long >:| Class Members | ||
|---|---|---|
| typedef float | Type_t | |
| struct qmcplusplus::Promote< float, short > |
Definition at line 323 of file TypeComputations.h.
 Collaboration diagram for Promote< float, short >:
Collaboration diagram for Promote< float, short >:| Class Members | ||
|---|---|---|
| typedef float | Type_t | |
| struct qmcplusplus::Promote< int, bool > |
Definition at line 223 of file TypeComputations.h.
 Collaboration diagram for Promote< int, bool >:
Collaboration diagram for Promote< int, bool >:| Class Members | ||
|---|---|---|
| typedef int | Type_t | |
| struct qmcplusplus::Promote< int, char > |
Definition at line 229 of file TypeComputations.h.
 Collaboration diagram for Promote< int, char >:
Collaboration diagram for Promote< int, char >:| Class Members | ||
|---|---|---|
| typedef int | Type_t | |
| struct qmcplusplus::Promote< int, double > |
Definition at line 259 of file TypeComputations.h.
 Collaboration diagram for Promote< int, double >:
Collaboration diagram for Promote< int, double >:| Class Members | ||
|---|---|---|
| typedef double | Type_t | |
| struct qmcplusplus::Promote< int, float > |
Definition at line 253 of file TypeComputations.h.
 Collaboration diagram for Promote< int, float >:
Collaboration diagram for Promote< int, float >:| Class Members | ||
|---|---|---|
| typedef float | Type_t | |
| struct qmcplusplus::Promote< int, int > |
Definition at line 241 of file TypeComputations.h.
 Collaboration diagram for Promote< int, int >:
Collaboration diagram for Promote< int, int >:| Class Members | ||
|---|---|---|
| typedef int | Type_t | |
| struct qmcplusplus::Promote< int, long > |
Definition at line 247 of file TypeComputations.h.
 Collaboration diagram for Promote< int, long >:
Collaboration diagram for Promote< int, long >:| Class Members | ||
|---|---|---|
| typedef long | Type_t | |
| struct qmcplusplus::Promote< int, short > |
Definition at line 235 of file TypeComputations.h.
 Collaboration diagram for Promote< int, short >:
Collaboration diagram for Promote< int, short >:| Class Members | ||
|---|---|---|
| typedef int | Type_t | |
| struct qmcplusplus::Promote< long, bool > |
Definition at line 267 of file TypeComputations.h.
 Collaboration diagram for Promote< long, bool >:
Collaboration diagram for Promote< long, bool >:| Class Members | ||
|---|---|---|
| typedef long | Type_t | |
| struct qmcplusplus::Promote< long, char > |
Definition at line 273 of file TypeComputations.h.
 Collaboration diagram for Promote< long, char >:
Collaboration diagram for Promote< long, char >:| Class Members | ||
|---|---|---|
| typedef long | Type_t | |
| struct qmcplusplus::Promote< long, double > |
Definition at line 303 of file TypeComputations.h.
 Collaboration diagram for Promote< long, double >:
Collaboration diagram for Promote< long, double >:| Class Members | ||
|---|---|---|
| typedef double | Type_t | |
| struct qmcplusplus::Promote< long, float > |
Definition at line 297 of file TypeComputations.h.
 Collaboration diagram for Promote< long, float >:
Collaboration diagram for Promote< long, float >:| Class Members | ||
|---|---|---|
| typedef float | Type_t | |
| struct qmcplusplus::Promote< long, int > |
Definition at line 285 of file TypeComputations.h.
 Collaboration diagram for Promote< long, int >:
Collaboration diagram for Promote< long, int >:| Class Members | ||
|---|---|---|
| typedef long | Type_t | |
| struct qmcplusplus::Promote< long, long > |
Definition at line 291 of file TypeComputations.h.
 Collaboration diagram for Promote< long, long >:
Collaboration diagram for Promote< long, long >:| Class Members | ||
|---|---|---|
| typedef long | Type_t | |
| struct qmcplusplus::Promote< long, short > |
Definition at line 279 of file TypeComputations.h.
 Collaboration diagram for Promote< long, short >:
Collaboration diagram for Promote< long, short >:| Class Members | ||
|---|---|---|
| typedef long | Type_t | |
| struct qmcplusplus::Promote< short, bool > |
Definition at line 179 of file TypeComputations.h.
 Collaboration diagram for Promote< short, bool >:
Collaboration diagram for Promote< short, bool >:| Class Members | ||
|---|---|---|
| typedef short | Type_t | |
| struct qmcplusplus::Promote< short, char > |
Definition at line 185 of file TypeComputations.h.
 Collaboration diagram for Promote< short, char >:
Collaboration diagram for Promote< short, char >:| Class Members | ||
|---|---|---|
| typedef short | Type_t | |
| struct qmcplusplus::Promote< short, double > |
Definition at line 215 of file TypeComputations.h.
 Collaboration diagram for Promote< short, double >:
Collaboration diagram for Promote< short, double >:| Class Members | ||
|---|---|---|
| typedef double | Type_t | |
| struct qmcplusplus::Promote< short, float > |
Definition at line 209 of file TypeComputations.h.
 Collaboration diagram for Promote< short, float >:
Collaboration diagram for Promote< short, float >:| Class Members | ||
|---|---|---|
| typedef float | Type_t | |
| struct qmcplusplus::Promote< short, int > |
Definition at line 197 of file TypeComputations.h.
 Collaboration diagram for Promote< short, int >:
Collaboration diagram for Promote< short, int >:| Class Members | ||
|---|---|---|
| typedef int | Type_t | |
| struct qmcplusplus::Promote< short, long > |
Definition at line 203 of file TypeComputations.h.
 Collaboration diagram for Promote< short, long >:
Collaboration diagram for Promote< short, long >:| Class Members | ||
|---|---|---|
| typedef long | Type_t | |
| struct qmcplusplus::Promote< short, short > |
Definition at line 191 of file TypeComputations.h.
 Collaboration diagram for Promote< short, short >:
Collaboration diagram for Promote< short, short >:| Class Members | ||
|---|---|---|
| typedef short | Type_t | |
| struct qmcplusplus::PSetsAndRefList |
Definition at line 79 of file test_ReferencePoints.cpp.
 Collaboration diagram for PSetsAndRefList:
Collaboration diagram for PSetsAndRefList:| Class Members | ||
|---|---|---|
| ParticleSetPool | ppool | |
| ParticleSet | pset | |
| ParticleSet | pset_ions | |
| RefVector< ParticleSet > | ref_psets | |
| class qmcplusplus::QMCTypes |
Definition at line 34 of file QMCTypes.h.
 Collaboration diagram for QMCTypes< Precision, DIM >:
Collaboration diagram for QMCTypes< Precision, DIM >:| Class Members | ||
|---|---|---|
| typedef complex< Precision > | ComplexType | |
| typedef TinyVector< ValueType, DIM > | GradType | |
| typedef TinyVector< RealType, DIM > | PosType | |
| typedef Precision | RealType | |
| typedef Tensor< RealType, DIM > | TensorType | |
| typedef RealType | ValueType | |
| struct qmcplusplus::RealAlias_impl |
Definition at line 38 of file complex_help.hpp.
 Collaboration diagram for RealAlias_impl< T, typename >:
Collaboration diagram for RealAlias_impl< T, typename >:| struct qmcplusplus::RealAlias_impl< T, IsComplex< T > > |
Definition at line 48 of file complex_help.hpp.
 Collaboration diagram for RealAlias_impl< T, IsComplex< T > >:
Collaboration diagram for RealAlias_impl< T, IsComplex< T > >:| Class Members | ||
|---|---|---|
| typedef typename value_type | value_type | |
| struct qmcplusplus::RealAlias_impl< T, IsReal< T > > |
Definition at line 42 of file complex_help.hpp.
 Collaboration diagram for RealAlias_impl< T, IsReal< T > >:
Collaboration diagram for RealAlias_impl< T, IsReal< T > >:| Class Members | ||
|---|---|---|
| typedef T | value_type | |
| struct qmcplusplus::RngReferenceVector< double > |
Definition at line 24 of file test_RandomForTest.cpp.
 Collaboration diagram for RngReferenceVector< double >:
Collaboration diagram for RngReferenceVector< double >:| Class Members | ||
|---|---|---|
| vector< double > | vec | |
| struct qmcplusplus::RngReferenceVector< float > |
Definition at line 30 of file test_RandomForTest.cpp.
 Collaboration diagram for RngReferenceVector< float >:
Collaboration diagram for RngReferenceVector< float >:| Class Members | ||
|---|---|---|
| vector< float > | vec | |
| struct qmcplusplus::RngReferenceVector< std::complex< double > > |
Definition at line 36 of file test_RandomForTest.cpp.
 Collaboration diagram for RngReferenceVector< std::complex< double > >:
Collaboration diagram for RngReferenceVector< std::complex< double > >:| Class Members | ||
|---|---|---|
| vector< complex< double > > | vec | |
| struct qmcplusplus::RngReferenceVector< std::complex< float > > |
Definition at line 42 of file test_RandomForTest.cpp.
 Collaboration diagram for RngReferenceVector< std::complex< float > >:
Collaboration diagram for RngReferenceVector< std::complex< float > >:| Class Members | ||
|---|---|---|
| vector< complex< float > > | vec | |
| class qmcplusplus::RPAFunctor |
| struct qmcplusplus::SplineStoragePrecision |
Definition at line 26 of file createBsplineReader.h.
 Collaboration diagram for SplineStoragePrecision< ST >:
Collaboration diagram for SplineStoragePrecision< ST >:| union qmcplusplus::StackKeyParam.__unnamed__ |
Definition at line 70 of file NewTimer.h.
 Collaboration diagram for StackKeyParam.__unnamed__:
Collaboration diagram for StackKeyParam.__unnamed__:| Class Members | ||
|---|---|---|
| long int | long_buckets[N] | |
| timer_id_t | short_buckets[sizeof(long int) *N/sizeof(timer_id_t)] | |
| struct qmcplusplus::SumCombine |
| struct qmcplusplus::TauParams |
Object to encapsulate appropriate tau derived parameters for a particular CoordsType specialization.
Definition at line 25 of file TauParams.hpp.
 Collaboration diagram for TauParams< RT, CT >:
Collaboration diagram for TauParams< RT, CT >:| struct qmcplusplus::TensorSoaContainer |
Definition at line 23 of file TensorSoaContainer.h.
 Collaboration diagram for TensorSoaContainer< T, D >:
Collaboration diagram for TensorSoaContainer< T, D >:| struct qmcplusplus::TimerIDName_t |
Definition at line 149 of file TimerManager.h.
 Collaboration diagram for TimerIDName_t< T >:
Collaboration diagram for TimerIDName_t< T >:| Class Members | ||
|---|---|---|
| T | id | |
| const string | name | |
| class qmcplusplus::TinyMatrix |
Definition at line 38 of file OhmmsTinyMeta.h.
 Collaboration diagram for TinyMatrix< T, D1, D2 >:
Collaboration diagram for TinyMatrix< T, D1, D2 >:| struct qmcplusplus::TreeCombine |
| struct qmcplusplus::TrinaryReturn |
Definition at line 454 of file TypeComputations.h.
 Collaboration diagram for TrinaryReturn< T1, T2, T3, Op >:
Collaboration diagram for TrinaryReturn< T1, T2, T3, Op >:| Class Members | ||
|---|---|---|
| typedef typename BinaryReturn< T2, T3, Op >::Type_t | Type_t | |
| struct qmcplusplus::TWFGrads |
Definition at line 22 of file TWFGrads.hpp.
 Collaboration diagram for TWFGrads< CT >:
Collaboration diagram for TWFGrads< CT >:| struct qmcplusplus::UnaryReturn |
Definition at line 64 of file TypeComputations.h.
 Collaboration diagram for UnaryReturn< T, Op >:
Collaboration diagram for UnaryReturn< T, Op >:| Class Members | ||
|---|---|---|
| typedef T | Type_t | |
| struct qmcplusplus::UnaryReturn< T, OpNot > |
Definition at line 261 of file OperatorTags.h.
 Collaboration diagram for UnaryReturn< T, OpNot >:
Collaboration diagram for UnaryReturn< T, OpNot >:| Class Members | ||
|---|---|---|
| typedef bool | Type_t | |
| struct qmcplusplus::UnaryReturn< T2, OpCast< T1 > > |
Definition at line 278 of file OperatorTags.h.
 Collaboration diagram for UnaryReturn< T2, OpCast< T1 > >:
Collaboration diagram for UnaryReturn< T2, OpCast< T1 > >:| Class Members | ||
|---|---|---|
| typedef T1 | Type_t | |
| struct qmcplusplus::UpdateEngineSelector |
Definition at line 36 of file DiracDeterminantBatched.h.
 Collaboration diagram for UpdateEngineSelector< PL, VT >:
Collaboration diagram for UpdateEngineSelector< PL, VT >:| struct qmcplusplus::UpdateEngineSelector< PlatformKind::OMPTARGET, VT > |
Definition at line 39 of file DiracDeterminantBatched.h.
 Collaboration diagram for UpdateEngineSelector< PlatformKind::OMPTARGET, VT >:
Collaboration diagram for UpdateEngineSelector< PlatformKind::OMPTARGET, VT >:| Class Members | ||
|---|---|---|
| typedef DelayedUpdateBatched< OMPTARGET, VT > | Engine | |
| struct qmcplusplus::ValGradLap |
Definition at line 445 of file CuspCorrectionConstruction.cpp.
 Collaboration diagram for ValGradLap:
Collaboration diagram for ValGradLap:| Class Members | ||
|---|---|---|
| GradType | grad | |
| ValueType | lap | |
| ValueType | val | |
| struct qmcplusplus::ValueAlias_impl |
Definition at line 63 of file complex_help.hpp.
 Collaboration diagram for ValueAlias_impl< TREAL, TREF, typename >:
Collaboration diagram for ValueAlias_impl< TREAL, TREF, typename >:| struct qmcplusplus::ValueAlias_impl< TREAL, TREF, IsComplex< TREF > > |
Definition at line 73 of file complex_help.hpp.
 Collaboration diagram for ValueAlias_impl< TREAL, TREF, IsComplex< TREF > >:
Collaboration diagram for ValueAlias_impl< TREAL, TREF, IsComplex< TREF > >:| Class Members | ||
|---|---|---|
| typedef complex< TREAL > | value_type | |
| struct qmcplusplus::ValueAlias_impl< TREAL, TREF, IsReal< TREF > > |
Definition at line 67 of file complex_help.hpp.
 Collaboration diagram for ValueAlias_impl< TREAL, TREF, IsReal< TREF > >:
Collaboration diagram for ValueAlias_impl< TREAL, TREF, IsReal< TREF > >:| Class Members | ||
|---|---|---|
| typedef TREAL | value_type | |
| struct qmcplusplus::WalkerLogState |
Helper struct holding data transferred from WalkerLogManager to WalkerLogCollector following input read.
Definition at line 31 of file WalkerLogCollector.h.
 Collaboration diagram for WalkerLogState:
Collaboration diagram for WalkerLogState:| Class Members | ||
|---|---|---|
| bool | logs_active | whether logs are active in the current driver |
| int | step_period | period between MC steps for data collection |
| bool | verbose | controls verbosity of log file writes |
| struct qmcplusplus::WaveFunctionTypes |
Consistent way to get the set of types used in the QMCWaveFunction module, without resorting to ifdef'd type aliases accessed through inheritance.
You can at least test all flavors without tricky scoping or recompiling. Determines type set for class through composition instead of inheritance.
Its somewhat question whether it is even worth have all these shortened type aliases Defined differently in different modules of the code, but it needs further study.
I would have liked to use QMCTypes but they do something dumb with handling complex which basically bakes in multiple builds for Real and Complex.
Definition at line 34 of file WaveFunctionTypes.hpp.
 Collaboration diagram for WaveFunctionTypes< VALUE, FP_VALUE >:
Collaboration diagram for WaveFunctionTypes< VALUE, FP_VALUE >:| Class Members | ||
|---|---|---|
| typedef complex< Real > | Complex | |
| typedef RealAlias< FullPrecValue > | FullPrecReal | |
| typedef FP_VALUE | FullPrecValue | |
| typedef TinyVector< Value, OHMMS_DIM > | Grad | |
| typedef Tensor< Value, OHMMS_DIM > | Hess | |
| typedef complex< FullPrecReal > | LogValue | |
| typedef FP_VALUE | PsiValue | |
| typedef RealAlias< Value > | Real | |
| typedef VALUE | Value | |
| struct qmcplusplus::WLog |
Basic data types for walker log data.
Definition at line 29 of file WalkerLogBuffer.h.
 Collaboration diagram for WLog:
Collaboration diagram for WLog:| Class Members | ||
|---|---|---|
| typedef complex< Real > | Comp | |
| typedef long | Int | |
| typedef Real | PsiVal | |
| typedef OHMMS_PRECISION_FULL | Real | |
| using aligned_allocator = Mallocator<T, ALIGN> |
Definition at line 33 of file aligned_allocator.hpp.
| using aligned_vector = std::vector<T, aligned_allocator<T> > |
Definition at line 35 of file aligned_allocator.hpp.
| using BasisSet_t = LCAOrbitalSet::basis_type |
Definition at line 118 of file LCAOrbitalBuilder.cpp.
| using ChronoClock = std::chrono::system_clock |
| typedef QMCTraits::ComplexType ComplexType |
Definition at line 33 of file test_DiracDeterminant.cpp.
Definition at line 39 of file PlatformSelector.hpp.
Definition at line 53 of file PlatformSelector.hpp.
| using CUDATypeMap = typename std::disjunction<OnTypesEqual<T, float, float>, OnTypesEqual<T, double, double>, OnTypesEqual<T, float*, float*>, OnTypesEqual<T, double*, double*>, OnTypesEqual<T, float**, float**>, OnTypesEqual<T, double**, double**>, OnTypesEqual<T, std::complex<double>, cuDoubleComplex>, OnTypesEqual<T, std::complex<float>, cuComplex>, OnTypesEqual<T, std::complex<double>*, cuDoubleComplex*>, OnTypesEqual<T, std::complex<float>**, cuComplex**>, OnTypesEqual<T, std::complex<double>**, cuDoubleComplex**>, OnTypesEqual<T, std::complex<float>*, cuComplex*>, OnTypesEqual<T, const std::complex<double>*, const cuDoubleComplex*>, OnTypesEqual<T, const std::complex<float>*, const cuComplex*>, OnTypesEqual<T, const std::complex<float>**, const cuComplex**>, OnTypesEqual<T, const std::complex<double>**, const cuDoubleComplex**>, OnTypesEqual<T, const std::complex<float>* const*, const cuComplex* const*>, OnTypesEqual<T, const std::complex<double>* const*, const cuDoubleComplex* const*>, default_type<void> >::type |
Definition at line 50 of file CUDATypeMapping.hpp.
| using DeviceAllocator = OffloadDeviceAllocator<T> |
Definition at line 60 of file DualAllocatorAliases.hpp.
| using DiracDet = DiracDeterminant<DelayedUpdate<QMCTraits::ValueType, QMCTraits::QTFull::ValueType> > |
Definition at line 36 of file test_TrialWaveFunction.cpp.
| using EstimatorInput = std::variant<std::monostate, MomentumDistributionInput, SpinDensityInput, OneBodyDensityMatricesInput, SelfHealingOverlapInput, MagnetizationDensityInput, PerParticleHamiltonianLoggerInput> |
Definition at line 52 of file EstimatorManagerInput.h.
| using EstimatorInputs = std::vector<EstimatorInput> |
Definition at line 53 of file EstimatorManagerInput.h.
| using FakeJasFunctor = FakeFunctor<OHMMS_PRECISION> |
Definition at line 21 of file test_J2_derivatives.cpp.
| using FakeTimer = TimerType<FakeChronoClock> |
Definition at line 235 of file NewTimer.h.
| using FakeTimerManager = TimerManager<FakeTimer> |
Definition at line 23 of file test_timer.cpp.
Definition at line 18 of file DMCRefEnergy.cpp.
Definition at line 18 of file HamiltonianRef.cpp.
Definition at line 24 of file test_DescentEngine.cpp.
| typedef TrialWaveFunction::GradType GradType |
Definition at line 21 of file LatticeGaussianProduct.cpp.
| typedef LRCoulombSingleton::GridType GridType |
Definition at line 30 of file DensityEstimator.cpp.
| using hipblasTypeMap = typename std::disjunction<OnTypesEqual<T, float, float>, OnTypesEqual<T, double, double>, OnTypesEqual<T, float*, float*>, OnTypesEqual<T, double*, double*>, OnTypesEqual<T, float**, float**>, OnTypesEqual<T, double**, double**>, OnTypesEqual<T, std::complex<double>, hipblasDoubleComplex>, OnTypesEqual<T, std::complex<float>, hipblasComplex>, OnTypesEqual<T, std::complex<double>*, hipblasDoubleComplex*>, OnTypesEqual<T, std::complex<float>**, hipblasComplex**>, OnTypesEqual<T, std::complex<double>**, hipblasDoubleComplex**>, OnTypesEqual<T, std::complex<float>*, hipblasComplex*>, OnTypesEqual<T, const std::complex<double>*, const hipblasDoubleComplex*>, OnTypesEqual<T, const std::complex<float>*, const hipblasComplex*>, OnTypesEqual<T, const std::complex<float>**, const hipblasComplex**>, OnTypesEqual<T, const std::complex<double>**, const hipblasDoubleComplex**>, OnTypesEqual<T, const std::complex<float>* const*, const hipblasComplex* const*>, OnTypesEqual<T, const std::complex<double>* const*, const hipblasDoubleComplex* const*>, default_type<void> >::type |
Definition at line 45 of file hipblasTypeMapping.hpp.
| using IndexType = QMCTraits::IndexType |
Definition at line 11 of file FSUtilities.h.
| typedef testing::ValidSpinDensityInput input |
Definition at line 131 of file test_SpinDensityNew.cpp.
| using IsComplex = std::enable_if_t<IsComplex_t<T>::value, bool> |
Definition at line 28 of file complex_help.hpp.
| using IsDualSpace = std::enable_if_t<qmc_allocator_traits<Allocator>::is_dual_space> |
Definition at line 52 of file allocator_traits.hpp.
| using IsHostSafe = std::enable_if_t<qmc_allocator_traits<Allocator>::is_host_accessible> |
Definition at line 46 of file allocator_traits.hpp.
| using IsNotHostSafe = std::enable_if_t<!qmc_allocator_traits<Allocator>::is_host_accessible> |
Definition at line 49 of file allocator_traits.hpp.
| using IsReal = std::enable_if_t<std::is_floating_point<T>::value, bool> |
Definition at line 35 of file complex_help.hpp.
| using IsReal_t = std::is_floating_point<T> |
Definition at line 32 of file complex_help.hpp.
| using LogComplexApprox = Catch::Detail::LogComplexApprox |
Definition at line 34 of file test_DiracMatrix.cpp.
| typedef TrialWaveFunction::LogValue LogValue |
Definition at line 25 of file test_cuSolverInverter.cpp.
Definition at line 29 of file DensityEstimator.cpp.
| typedef Walker< QMCTraits, PtclOnLatticeTraits > MCPWalker |
Definition at line 31 of file test_walker.cpp.
| typedef LRHandlerBase::mRealType mRealType |
Definition at line 21 of file test_ewald3d.cpp.
| using NewTimer = TimerType<ChronoClock> |
Definition at line 234 of file NewTimer.h.
| using OffloadAllocator = OMPallocator<T, aligned_allocator<T> > |
Definition at line 23 of file OffloadAlignedAllocators.hpp.
| using OffloadDeviceAllocator = aligned_allocator<T> |
Definition at line 30 of file OffloadAlignedAllocators.hpp.
| using OffloadPinnedAllocator = OMPallocator<T, PinnedAlignedAllocator<T> > |
Definition at line 33 of file test_dual_allocators_ohmms_containers.cpp.
| using OffloadPinnedMatrix = Matrix<T, OffloadPinnedAllocator<T> > |
Definition at line 40 of file benchmark_DiracMatrixComputeCUDA.cpp.
| using OffloadPinnedVector = Vector<T, OffloadPinnedAllocator<T> > |
Definition at line 42 of file benchmark_DiracMatrixComputeCUDA.cpp.
| using OffloadVector = Vector<DT, OffloadPinnedAllocator<DT> > |
Definition at line 35 of file test_LCAO_diamondC_2x1x1.cpp.
Definition at line 24 of file OptimizableObject.h.
| using ParseCaseVector_t = vector<ParseCase> |
Definition at line 47 of file test_parser.cpp.
| using PinnedAlignedAllocator = aligned_allocator<T, ALIGN> |
Definition at line 43 of file PinnedAllocator.h.
| using PinnedAllocator = std::allocator<T> |
The fact that the pinned allocators are not always pinned hurts readability elsewhere.
Definition at line 34 of file PinnedAllocator.h.
| using PinnedDualAllocator = OffloadPinnedAllocator<T> |
Definition at line 58 of file DualAllocatorAliases.hpp.
| typedef SkParserScalarDat::PosType PosType |
Definition at line 18 of file test_kcontainer.cpp.
| using pRealType = qmcplusplus::LRHandlerBase::pRealType |
Definition at line 21 of file test_lrhandler.cpp.
| typedef TrialWaveFunction::PsiValue PsiValue |
Definition at line 25 of file SlaterDet.cpp.
Definition at line 33 of file test_bare_kinetic.cpp.
| using QuantumNumberType = TinyVector<int, 4> |
Definition at line 37 of file OrbitalSetTraits.h.
Definition at line 31 of file DensityEstimator.cpp.
| using RandomGenerator = StdRandom<OHMMS_PRECISION_FULL> |
Definition at line 55 of file RandomGenerator.h.
| typedef QMCTraits::RealType Real |
Definition at line 26 of file ForceBase.cpp.
| typedef OHMMS_PRECISION real_type |
Definition at line 23 of file test_cartesian_tensor.cpp.
| using RealAlias = typename RealAlias_impl<T>::value_type |
If you have a function templated on a value that can be real or complex and you need to get the base Real type if its complex or just the real.
If you try to do this on anything but a fp or a std::complex<fp> you will get a compilation error.
Definition at line 60 of file complex_help.hpp.
| typedef SkParserScalarDat::RealType RealType |
Definition at line 22 of file test_min_oned.cpp.
| using RefVector = std::vector<std::reference_wrapper<T> > |
Definition at line 32 of file template_types.hpp.
| using Return_t = BareKineticEnergy::Return_t |
Definition at line 29 of file BareKineticEnergy.cpp.
| using RngValueType = typename std::disjunction<OnTypesEqual<T, float, float>, OnTypesEqual<T, double, double>, OnTypesEqual<T, std::complex<float>, float>, OnTypesEqual<T, std::complex<double>, double>, default_type<void> >::type |
Get the right type for rng in case of complex T.
Probably a more elegant way to do this especially in c++17
Definition at line 31 of file makeRngSpdMatrix.hpp.
| using ScalarEstimatorInput = std::variant<std::monostate, LocalEnergyInput, CSLocalEnergyInput, RMCLocalEnergyInput> |
Definition at line 60 of file EstimatorManagerInput.h.
| using ScalarEstimatorInputs = std::vector<ScalarEstimatorInput> |
Definition at line 61 of file EstimatorManagerInput.h.
| using ScopedFakeTimer = ScopeGuard<FakeTimer> |
Definition at line 258 of file NewTimer.h.
| using ScopedTimer = ScopeGuard<NewTimer> |
Definition at line 257 of file NewTimer.h.
| using SPOMap = SPOSet::SPOMap |
Definition at line 48 of file EstimatorManagerNew.cpp.
| using StackKey = StackKeyParam<2> |
Definition at line 129 of file NewTimer.h.
| using StdComp = std::complex<double> |
Definition at line 221 of file test_cuBLAS_LU.cpp.
| using timer_id_t = unsigned char |
Definition at line 56 of file NewTimer.h.
| using TimerList_t = TimerList<NewTimer> |
Definition at line 177 of file TimerManager.h.
| using TimerNameList_t = std::vector<TimerIDName_t<T> > |
Definition at line 156 of file TimerManager.h.
Definition at line 50 of file TraceManager.h.
| using TraceInt = long |
Definition at line 48 of file TraceManager.h.
| using TraceReal = OHMMS_PRECISION |
Definition at line 49 of file TraceManager.h.
| using UnpinnedDualAllocator = OffloadAllocator<T> |
Definition at line 56 of file DualAllocatorAliases.hpp.
| using UPtr = std::unique_ptr<T> |
Definition at line 35 of file template_types.hpp.
| using UPtrVector = std::vector<std::unique_ptr<T> > |
Definition at line 38 of file template_types.hpp.
| using Value_t = DensityMatrices1B::Value_t |
Definition at line 1088 of file DensityMatrices1B.cpp.
Definition at line 28 of file ObservableHelper.h.
| using ValueAlias = typename ValueAlias_impl<TREAL, TREF>::value_type |
If you need to make a value type of a given precision based on a reference value type set the desired POD float point type as TREAL and set the reference type as TREF.
If TREF is real/complex, the generated Value type is real/complex.
Definition at line 83 of file complex_help.hpp.
Definition at line 20 of file LatticeGaussianProduct.cpp.
| using ValueVector = OrbitalSetTraits<ValueType>::ValueVector |
Definition at line 129 of file CuspCorrectionConstruction.h.
| typedef TinyVector< OHMMS_PRECISION, 3 > vec_t |
Definition at line 24 of file test_CrystalLattice.cpp.
| typedef WalkerProperties::Indexes WP |
Definition at line 34 of file ParticleSet.cpp.
| anonymous enum |
enumeration to classify a CrystalLattice
Use std::bitset<3> for all the dimension
| Enumerator | |
|---|---|
| SUPERCELL_OPEN | nnn |
| SUPERCELL_WIRE | nnp |
| SUPERCELL_SLAB | npp |
| SUPERCELL_BULK | ppp |
| SOA_OFFSET | const to differentiate AoS and SoA |
Definition at line 37 of file CrystalLattice.h.
| anonymous enum |
enumeration for DTD_BConds specialization
G = general cell with image-cell checks S = special treatment of a general cell with Wigner-cell radius == Simulation cell O = orthogonal cell X = exhaustive search (reference implementation)
| Enumerator | |
|---|---|
| PPPG | |
| PPPS | |
| PPPO | |
| PPPX | |
| PPNG | |
| PPNS | |
| PPNO | |
| PPNX | |
Definition at line 28 of file LatticeAnalyzer.h.
| anonymous enum |
| Enumerator | |
|---|---|
| q_n | |
| q_l | |
| q_m | |
| q_s | |
Definition at line 39 of file OrbitalSetTraits.h.
| anonymous enum |
Tmove options.
| Enumerator | |
|---|---|
| TMOVE_OFF | |
| TMOVE_V0 | |
| TMOVE_V1 | |
| TMOVE_V3 | |
Definition at line 29 of file NonLocalTOperator.h.
| anonymous enum |
enumeration for EstimatorManagerBase.Options
| Enumerator | |
|---|---|
| COLLECT | |
| MANAGE | |
| RECORD | |
| POSTIRECV | |
| APPEND | |
Definition at line 46 of file EstimatorManagerBase.cpp.
| anonymous enum |
enum to yes/no options saved in sParam
| Enumerator | |
|---|---|
| COMBOPT | |
| USETAUOPT | |
| MIXDMCOPT | |
| DUMMYOPT | |
Definition at line 26 of file SFNBranch.cpp.
| anonymous enum |
enum to yes/no options saved in sParam
| Enumerator | |
|---|---|
| COMBOPT | |
| USETAUOPT | |
| MIXDMCOPT | |
| DUMMYOPT | |
Definition at line 37 of file SimpleFixedNodeBranch.cpp.
|
strong |
| Enumerator | |
|---|---|
| POS | |
| POS_SPIN | |
Definition at line 24 of file MCCoords.hpp.
|
strong |
data locality with respect to walker buffer
| Enumerator | |
|---|---|
| process | |
| rank | |
| crowd | |
| queue | |
| walker | |
Definition at line 19 of file DataLocality.h.
|
strong |
determinant matrix inverter select
| Enumerator | |
|---|---|
| HOST | |
| ACCEL | |
Definition at line 27 of file DiracDeterminantBase.h.
| enum DMC_MPI_Timers |
| Enumerator | |
|---|---|
| DMC_MPI_branch | |
| DMC_MPI_imbalance | |
| DMC_MPI_prebalance | |
| DMC_MPI_copyWalkers | |
| DMC_MPI_allreduce | |
| DMC_MPI_loadbalance | |
| DMC_MPI_send | |
| DMC_MPI_recv | |
Definition at line 29 of file WalkerControlMPI.cpp.
|
strong |
DMCRefEnergy schemes.
| Enumerator | |
|---|---|
| UNLIMITED_HISTORY | |
| LIMITED_HISTORY | |
Definition at line 19 of file DMCRefEnergyScheme.h.
| enum DMCTimers |
| Enumerator | |
|---|---|
| DMC_buffer | |
| DMC_movePbyP | |
| DMC_hamiltonian | |
| DMC_collectables | |
| DMC_tmoves | |
Definition at line 40 of file DMCUpdatePbyP.h.
| enum DMTimers |
| Enumerator | |
|---|---|
| DM_eval | |
| DM_gen_samples | |
| DM_gen_sample_basis | |
| DM_gen_sample_ratios | |
| DM_gen_particle_basis | |
| DM_matrix_products | |
| DM_accumulate | |
Definition at line 28 of file DensityMatrices1B.cpp.
|
strong |
| Enumerator | |
|---|---|
| ALL_OFF | |
| CHECKGL_AFTER_LOAD | |
| CHECKGL_AFTER_MOVES | |
| CHECKGL_AFTER_TMOVE | |
Definition at line 20 of file DriverDebugChecks.h.
|
strong |
| Enumerator | |
|---|---|
| ALL_OFF | |
| NEED_FULL_TABLE_ANYTIME | whether full table needs to be ready at anytime or not during PbyP Optimization can be implemented during forward PbyP move when the full table is not needed all the time. DT consumers should know if full table is needed or not and request via addTable. |
| NEED_VP_FULL_TABLE_ON_HOST | For distance tables of virtual particle (VP) sets constructed based on this table, whether full table is needed on host The corresponding DT of VP need to set MW_EVALUATE_RESULT_NO_TRANSFER_TO_HOST accordingly. |
| NEED_TEMP_DATA_ON_HOST | whether temporary data set on the host is updated or not when a move is proposed. Considering transferring data from accelerator to host is relatively expensive, only request this when data on host is needed for unoptimized code path. This flag affects three subroutines mw_move, mw_updatePartial, mw_finalizePbyP in DistanceTable. |
| MW_EVALUATE_RESULT_NO_TRANSFER_TO_HOST | skip data transfer back to host after mw_evalaute full distance table. this optimization can be used for distance table consumed directly on the device without copying back to the host. |
| NEED_FULL_TABLE_ON_HOST_AFTER_DONEPBYP | whether full table needs to be ready at anytime or not after donePbyP Optimization can be implemented during forward PbyP move when the full table is not needed all the time. DT consumers should know if full table is needed or not and request via addTable. |
Definition at line 20 of file DTModes.h.
|
strong |
enumerator for DynamicCoordinates kinds
| Enumerator | |
|---|---|
| DC_POS | |
| DC_POS_OFFLOAD | |
Definition at line 29 of file DynamicCoordinates.h.
|
strong |
| Enumerator | |
|---|---|
| NONE | |
| QUARTIC | |
| RESCALE | |
| LINEMIN | |
| ONESHIFTONLY | |
| ADAPTIVE | |
| DESCENT | |
| HYBRID | |
| GRADIENT_TEST | |
Definition at line 17 of file OptimizerTypes.h.
|
strong |
| Enumerator | |
|---|---|
| CPU | |
| OMPTARGET | |
| CUDA | |
| SYCL | |
Definition at line 19 of file PlatformKinds.hpp.
|
strong |
| enum PSetTimers |
| Enumerator | |
|---|---|
| PS_newpos | |
| PS_donePbyP | |
| PS_accept | |
| PS_loadWalker | |
| PS_update | |
| PS_dt_move | |
| PS_mw_copy | |
Definition at line 36 of file ParticleSet.cpp.
| enum QMCModeEnum |
enum to set the bit to determine the QMC mode
Can't be a scoped enum, unsafe out in qmcplusplus scope
Definition at line 26 of file DriverTraits.h.
|
strong |
enum for QMC Run Type
| Enumerator | |
|---|---|
| DUMMY | dummy |
| VMC | VMC type: vmc, vmc-ptcl, vmc-multiple, vmc-ptcl-multiple. |
| CSVMC | |
| DMC | DMC type: dmc, dmc-ptcl. |
| RMC | RMC type: rmc, rmc-ptcl. |
| VMC_OPT | Optimization with vmc blocks |
| LINEAR_OPTIMIZE | |
| WF_TEST | |
| VMC_BATCH | |
| DMC_BATCH | |
| LINEAR_OPTIMIZE_BATCH | |
Definition at line 7 of file DriverTraits.h.
|
strong |
| Enumerator | |
|---|---|
| CPU_OMPTARGET | |
| CPU_OMPTARGET_CUDA | |
| CPU_OMPTARGET_SYCL | |
Definition at line 24 of file PlatformSelector.hpp.
|
strong |
| enum SoAFields3D |
| Enumerator | |
|---|---|
| VAL | |
| GRAD0 | |
| GRAD1 | |
| GRAD2 | |
| HESS00 | |
| HESS01 | |
| HESS02 | |
| HESS11 | |
| HESS12 | |
| HESS22 | |
| LAPL | |
| NUM_FIELDS | |
Definition at line 18 of file contraction_helper.hpp.
| enum SODMCTimers |
| Enumerator | |
|---|---|
| SODMC_buffer | |
| SODMC_movePbyP | |
| SODMC_hamiltonian | |
| SODMC_collectables | |
| SODMC_tmoves | |
Definition at line 37 of file SODMCUpdatePbyP.h.
|
strong |
| Enumerator | |
|---|---|
| VALUE1 | |
| VALUE2 | |
Definition at line 31 of file test_InputSection.cpp.
|
strong |
| Enumerator | |
|---|---|
| VALUE1 | |
| VALUE2 | |
Definition at line 37 of file test_InputSection.cpp.
| enum TestTimer |
| Enumerator | |
|---|---|
| MyTimer1 | |
| MyTimer2 | |
Definition at line 390 of file test_timer.cpp.
| enum timer_levels |
| Enumerator | |
|---|---|
| timer_level_none | |
| timer_level_coarse | |
| timer_level_medium | |
| timer_level_fine | |
| num_timer_levels | |
Definition at line 42 of file NewTimer.h.
| enum TimerEnum |
| Enumerator | |
|---|---|
| V_TIMER | |
| VGL_TIMER | |
| ACCEPT_TIMER | |
| NL_TIMER | |
| RECOMPUTE_TIMER | |
| BUFFER_TIMER | |
| DERIVS_TIMER | |
| PREPAREGROUP_TIMER | |
| TIMER_SKIP | |
Definition at line 31 of file TrialWaveFunction.cpp.
| enum WC_Timers |
| Enumerator | |
|---|---|
| WC_branch | |
| WC_imbalance | |
| WC_prebalance | |
| WC_copyWalkers | |
| WC_recomputing | |
| WC_allreduce | |
| WC_loadbalance | |
| WC_send | |
| WC_recv | |
Definition at line 34 of file WalkerControl.cpp.
|
inline |
Definition at line 88 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by JeeIOrbitalSoA< FT >::acceptMove(), LRBreakup< BreakupBasis >::AddKToList(), EinsplineSetBuilder::AnalyzeTwists2(), BareKineticEnergy::BareKineticEnergy(), KContainer::BuildKLists(), ECPComponentBuilder::buildLocal(), OneBodyDensityMatrices::calcDensity(), OneBodyDensityMatrices::calcDensityDrift(), OneBodyDensityMatrices::calcDensityDriftWithSpin(), OneBodyDensityMatrices::calcDensityWithSpin(), LatticeAnalyzer< T, 3 >::calcSimulationCellRadius(), BsplineReader::check_twists(), GKIntegration< F, GKRule >::CheckError(), DiracDeterminantBase::checkG(), WaveFunctionTester::checkGradients(), EinsplineSetBuilder::CheckLattice(), QMCUpdateBase::checkLogAndGL(), QMCDriverNew::checkLogAndGL(), Quadrature3D< T >::CheckQuadratureRule(), Quadrature3D< T >::CheckQuadratureRuleReal(), WalkerLogCollector::collect(), ACForce::compute_regularizer_f(), QMCCostFunctionBase::computedCost(), DescentEngine::computeFinalizationUncertainties(), FiniteDifference::computeFiniteDiffRichardson(), QMCCostFunction::correlatedSampling(), QMCCostFunctionBatched::correlatedSampling(), CoulombPBCAA::CoulombPBCAA(), SPOSetInfo::count_degeneracies(), SlaterDetBuilder::createMSDFast(), QMCGaussianParserBase::createMultiDeterminantSet(), FreeOrbitalBuilder::createSPOSetFromXML(), ECPComponentBuilder::createVrWithBasisGroup(), DensityMatrices1B::density_drift(), DensityMatrices1B::density_only(), RPABFeeBreakup< T >::Dlindhard(), RPABFeeBreakup< T >::Dlindhardp(), kSpaceJastrow::Equivalent(), EwaldHandlerQuasi2D::evaluate_slab(), LocalECPotential::evaluate_sp(), BareKineticEnergy::evaluate_sp(), CoulombPBCAB::evaluate_sp(), CoulombPBCAA::evaluate_sp(), CoulombPotential< T >::evaluate_spAA(), CoulombPotential< T >::evaluate_spAB(), SphericalTensor< T, Point_t, Tensor_t, GGG_t >::evaluateAll(), PolynomialFunctor3D::evaluateDerivatives(), NonLocalECPotential::evaluateImpl(), NonLocalECPComponent::evaluateOneBodyOpMatrixdRContribution(), NonLocalECPComponent::evaluateOneWithForces(), SoaSphericalTensor< ST >::evaluateVGL_impl(), SphericalTensor< T, Point_t, Tensor_t, GGG_t >::evaluateWithHessian(), evalX(), qmcplusplus::ewaldref::ewaldEnergy(), qmcplusplus::ewaldref::ewaldSum(), QMCMain::executeCMCSection(), RadialOrbitalSetBuilder< COT >::find_cutoff(), find_minimum(), generateCuspInfo(), GamesAsciiParser::getCSF(), WalkerReconfiguration::getIndexPermutation(), GamesAsciiParser::getORMAS(), getScore(), GKIntegration< F, GKRule >::GKGeneral(), QMCCostFunction::GradCost(), QMCCostFunctionBatched::GradCost(), qmcplusplus::ewaldref::gridSum(), CenterGrid::Init(), MPC::init_f_G(), MPC::init_gvecs(), CoulombPBCAB::initBreakup(), SpaceGrid::initialize_rectilinear(), InitMolecularSystem::initWithVolume(), GKIntegration< F, GKRule >::Integrate(), QMCFixedSampleLinearOptimizeBatched::is_best_cost(), LatticeAnalyzer< T, 3 >::isDiagonalOnly(), LatticeAnalyzer< T, 2 >::isDiagonalOnly(), ChannelPotential::jl(), LegendrePlm(), LCAOrbitalBuilder::LoadFullCoefsFromH5(), qmcplusplus::ewaldref::madelungSum(), mag(), DensityMatrices1B::match(), NonLocalECPotential::mw_evaluateImpl(), SPOSetInputInfo::occupy_energies(), QMCFixedSampleLinearOptimizeBatched::one_shift_run(), operator!=(), EnergyOrder::operator()(), FnFabs::operator()(), kSpaceJastrow::operator()(), KPoint::operator<(), KPoint::operator==(), operator==(), operator==(), CrystalLattice< ST, 3 >::outOfBound(), GaussianFCHKParser::parse(), ECPComponentBuilder::parseCasino(), parseGridInput(), lup::permute_max_diagonal(), PolynomialFunctor3D::PolynomialFunctor3D(), QMCFixedSampleLinearOptimizeBatched::previous_linear_methods_run(), LCAOrbitalBuilder::putPBCFromH5(), Quadrature3D< T >::Quadrature3D(), QuinticSplineSolve(), SkParserScalarDat::read_sk_file(), SlaterDetBuilder::readDetList(), SlaterDetBuilder::readDetListH5(), EinsplineSetBuilder::ReadGvectors_ESHDF(), SplineSetReader< typename SA::SplineBase >::readOneOrbitalCoefs(), EinsplineSetBuilder::ReadOrbitalInfo_ESHDF(), WalkerReconfigurationMPI::recvWalkers(), CrystalLattice< ST, 3 >::reset(), PolynomialFunctor3D::reset_gamma(), PolynomialFunctor3D::resize(), CenterGrid::ReverseMap(), QMCFixedSampleLinearOptimize::run(), GradientTest::run(), WaveFunctionTester::runBasicTest(), WalkerReconfigurationMPI::sendWalkers(), SimCellRad(), NESpaceGrid< REAL >::someMoreAxisGridStuff(), WalkerReconfigurationMPI::swapWalkers(), TEST_CASE(), SHOSet::test_derivatives(), SHOSet::test_overlap(), DensityMatrices1B::test_overlap(), BackflowTransformation::testDeriv(), DiracDeterminantWithBackflow::testDerivFjj(), OneBodyDensityMatricesTests< T >::testGenerateSamplesForSpinor(), DiracDeterminantWithBackflow::testGGG(), testJastrow(), PWBasis::trimforecut(), EinsplineSetBuilder::TwistPair(), DescentEngine::updateParameters(), EnergyDensityEstimator::write_nonzero_domains(), and OrbitalImages::write_orbital_xsf().
|
inline |
Definition at line 1435 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1091 of file DensityMatrices1B.cpp.
References CombinedTraceSample< T >::combine(), TraceSample< T >::sample, and Vector< T, Alloc >::size().
Referenced by DensityMatrices1B::get_energies().
|
inline |
Definition at line 1104 of file DensityMatrices1B.cpp.
References CombinedTraceSample< T >::combine(), TraceSample< T >::sample, and Vector< T, Alloc >::size().
Referenced by DensityMatrices1B::get_energies().
|
inline |
Definition at line 1115 of file DensityMatrices1B.cpp.
References if(), real(), TraceSample< T >::sample, and Vector< T, Alloc >::size().
|
inline |
Definition at line 34 of file IteratorUtility.h.
Referenced by SkEstimator::evaluate(), and SkAllEstimator::evaluate().
| void qmcplusplus::accumulateFromPsets | ( | int | ncrowds, |
| SpinDensityNew & | sdn, | ||
| UPtrVector< OperatorEstBase > & | crowd_sdns | ||
| ) |
Definition at line 51 of file test_SpinDensityNew.cpp.
References SpinDensityNew::accumulate(), pset, sdn, SpinDensityNew::spawnCrowdClone(), and qmcplusplus::hdf::walkers.
|
inline |
Definition at line 32 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by NESpaceGrid< REAL >::accumulate(), LatticeAnalyzer< T, 2 >::calcSimulationCellRadius(), LatticeAnalyzer< T, 3 >::calcSolidAngles(), LatticeAnalyzer< T, 2 >::calcSolidAngles(), cart2sph(), SpaceGrid::check_grid(), NESpaceGrid< REAL >::check_grid(), CubicFormula(), derivYlmSpherical(), SpaceGrid::evaluate(), FnArcCos::operator()(), and Quadrature3D< T >::Quadrature3D().
|
inline |
Definition at line 1379 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
| oh qmcplusplus::addProperty | ( | propertyFloat | , |
| "propertyFloat" | , | ||
| hFile | |||
| ) |
| oh qmcplusplus::addProperty | ( | propertyTensor | , |
| "propertyTensor" | , | ||
| hFile | |||
| ) |
| oh qmcplusplus::addProperty | ( | propertyMatrix | , |
| "propertyMatrix" | , | ||
| hFile | |||
| ) |
| oh qmcplusplus::addProperty | ( | propertyTensor | , |
| "propertyTinyVector" | , | ||
| hFile | |||
| ) |
| oh qmcplusplus::addProperty | ( | propertyVector | , |
| "propertyVector" | , | ||
| hFile | |||
| ) |
| oh qmcplusplus::addProperty | ( | propertyVectorTinyVector | , |
| "propertyVectorTinyVector" | , | ||
| hFile | |||
| ) |
|
inline |
Definition at line 71 of file OutputManager.h.
References InfoStream::getStream(), and infoDebug.
Referenced by ResourceCollection::addResource(), VMCBatched::run(), and DMCBatched::run().
|
inline |
Definition at line 67 of file OutputManager.h.
References InfoStream::getStream(), and infoError.
Referenced by PWOrbitalSet::addVector(), PWRealOrbitalSet::addVector(), EinsplineSetBuilder::AnalyzeTwists2(), WalkerControlBase::applyNmaxNmin(), JastrowBuilder::build_eeI(), LatticeGaussianProductBuilder::buildComponent(), ExampleHeBuilder::buildComponent(), kSpaceJastrowBuilder::buildComponent(), RadialJastrowBuilder::buildComponent(), ECPComponentBuilder::buildL2(), ECPComponentBuilder::buildSemiLocalAndLocal(), EinsplineSetBuilder::CheckLattice(), Quadrature3D< T >::CheckQuadratureRule(), Quadrature3D< T >::CheckQuadratureRuleReal(), OneDimGridFactory::createGrid(), PWOrbitalSetBuilder::createPW(), PWOrbitalSetBuilder::createPWBasis(), WalkerControlMPI::determineNewWalkerPopulation(), WalkerControl::determineNewWalkerPopulation(), ReportEngine::error(), QMCMain::execute(), ExpFitClass< M >::Fit(), ExpFitClass< M >::FitCusp(), PWOrbitalSetBuilder::getH5(), XMLParticleParser::getPtclAttrib(), RadialJastrowBuilder::guardAgainstOBC(), RadialJastrowBuilder::guardAgainstPBC(), CSUpdateBase::initCSWalkers(), HybridRepSetReader< SA >::initialize_hybridrep_atomic_centers(), main(), EinsplineSetBuilder::OccupyBands(), ECPComponentBuilder::parseCasino(), PolynomialFunctor3D::PolynomialFunctor3D(), QMCAppBase::pushDocument(), HDFWalkerInput_0_4::put(), HDFWalkerInput_0_4::read_hdf5(), HDFWalkerInput_0_4::read_hdf5_scatter(), HDFWalkerInput_0_4::read_phdf5(), EinsplineSetBuilder::ReadOrbitalInfo(), EinsplineSetBuilder::ReadOrbitalInfo_ESHDF(), PolynomialFunctor3D::reset_gamma(), MCWalkerConfiguration::resetWalkerProperty(), BasisSetBase< T >::resize(), WalkerControlMPI::swapWalkersSimple(), TEST_CASE(), ECPotentialBuilder::useSimpleTableFormat(), and ECPotentialBuilder::useXmlFormat().
|
inline |
Definition at line 65 of file OutputManager.h.
References InfoStream::getStream(), and infoLog.
Referenced by NESpaceGrid< REAL >::accumulate(), EstimatorManagerBase::add(), CoulombPBCAB::add(), TrialWaveFunction::addComponent(), HamiltonianFactory::addCoulombPotential(), WaveFunctionPool::addFactory(), HamiltonianFactory::addForceHam(), Backflow_eI_spin< FT >::addFunc(), RadialOrbitalSetBuilder< COT >::addGridH5(), EstimatorManagerNew::addMainEstimator(), ForwardWalking::addObservables(), QMCHamiltonian::addObservables(), BackflowBuilder::addOneBody(), SymmetryGroup::addOperator(), QMCHamiltonian::addOperator(), QMCHamiltonian::addOperatorType(), BackflowBuilder::addRPA(), ParticleSet::addTable(), BackflowBuilder::addTwoBody(), QMCDriver::addWalkers(), RMCUpdatePbyPWithDrift::advanceWalkersRMC(), RMCUpdateAllWithDrift::advanceWalkersRMC(), EinsplineSetBuilder::AnalyzeTwists2(), SPOSet::basic_report(), SimpleFixedNodeBranch::branch(), TraceManager::buffer_sample(), HamiltonianFactory::build(), BackflowBuilder::buildBackflowTransformation(), SymmetryBuilder::buildByHand(), AGPDeterminantBuilder::buildComponent(), kSpaceJastrowBuilder::buildComponent(), SlaterDetBuilder::buildComponent(), JastrowBuilder::buildkSpace(), ECPComponentBuilder::buildL2(), ECPComponentBuilder::buildLocal(), RotatedSPOs::buildOptVariables(), RPAJastrow::buildOrbital(), ECPComponentBuilder::buildSemiLocalAndLocal(), ECPComponentBuilder::buildSO(), WaveFunctionFactory::buildTWF(), TraceManager::check_clones(), JeeIOrbitalSoA< FT >::check_complete(), SpaceGrid::check_grid(), NESpaceGrid< REAL >::check_grid(), BsplineReader::check_twists(), WalkerLogCollector::checkBuffers(), WalkerLogManager::checkCollectors(), QMCCostFunction::checkConfigurations(), QMCCostFunctionBatched::checkConfigurations(), TraceSamples< std::complex< TraceReal > >::checkout_array(), SimpleFixedNodeBranch::checkParameters(), MagnetizationDensityInput::MagnetizationDensityInputSection::checkParticularValidity(), PWParameterSet::checkVersion(), TraceManager::close_file(), WalkerLogManager::closeFile(), WalkerLogManager::closeHDFFile(), SimpleFixedNodeBranch::collect(), TraceBuffer< TraceInt >::collect_sample(), DensityMatrices1B::compare(), DescentEngine::computeFinalizationUncertainties(), CoulombPBCAA::CoulombPBCAA(), CoulombPBCAB::CoulombPBCAB(), HybridRepSetReader< SA >::create_atomic_centers_Gspace(), SplineC2R< ST >::create_spline(), SplineR2R< ST >::create_spline(), SplineC2C< ST >::create_spline(), SplineC2COMPTarget< ST >::create_spline(), SplineC2ROMPTarget< ST >::create_spline(), HybridRepSetReader< SA >::create_spline_set(), SplineSetReader< typename SA::SplineBase >::create_spline_set(), AGPDeterminantBuilder::createAGP(), AOBasisBuilder< COT >::createAOSet(), AOBasisBuilder< COT >::createAOSetH5(), LCAOrbitalBuilder::createBasisSetH5(), CountingJastrowBuilder::createCJ(), MultiDiracDeterminant::createDetData(), ECPComponentBuilder::createGrid(), RadialJastrowBuilder::createJ1(), SlaterDetBuilder::createMSDFast(), QMCGaussianParserBase::createMultiDeterminantSetFromH5(), PWOrbitalSetBuilder::createPW(), PWOrbitalSetBuilder::createPWBasis(), QMCDriverFactory::createQMCDriver(), ParticleSet::createSK(), SplineSetReader< typename SA::SplineBase >::createSplineDataSpaceLookforDumpFile(), SHOSetBuilder::createSPOSet(), SPOSetBuilder::createSPOSet(), SPOSetBuilderFactory::createSPOSetBuilder(), FreeOrbitalBuilder::createSPOSetFromXML(), LCAOrbitalBuilder::createSPOSetFromXML(), LCAOSpinorBuilder::createSPOSetFromXML(), SHOSetBuilder::createSPOSetFromXML(), EinsplineSetBuilder::createSPOSetFromXML(), ECPComponentBuilder::createVrWithBasisGroup(), createWalkerController(), ECPComponentBuilder::doBreakUp(), QMCGaussianParserBase::dump(), VMCBatched::enable_sample_collection(), EstimatorManagerBaseTest::EstimatorManagerBaseTest(), EstimatorManagerNew::EstimatorManagerNew(), EstimatorManagerNewTest::EstimatorManagerNewTest(), CoulombPBCAB::evalConsts(), StressPBC::evalConsts_AA(), EnergyDensityEstimator::evaluate(), LatticeDeviationEstimator::evaluate(), SpaceGrid::evaluate(), OrbitalImages::evaluate(), SHOSet::evaluate_check(), DensityMatrices1B::evaluate_matrix(), LocalECPotential::evaluate_sp(), BareKineticEnergy::evaluate_sp(), CoulombPBCAB::evaluate_sp(), CoulombPBCAA::evaluate_sp(), CoulombPotential< T >::evaluate_spAA(), CoulombPotential< T >::evaluate_spAB(), CountingJastrow< RegionType >::evaluateDerivatives(), CountingJastrow< RegionType >::evaluateExponents(), CountingJastrow< RegionType >::evaluateExponents_print(), NonLocalECPotential::evaluateImpl(), CountingJastrow< RegionType >::evaluateTempExponents(), CountingJastrow< RegionType >::evaluateTempExponents_print(), EwaldHandler2D::EwaldHandler2D(), EwaldHandlerQuasi2D::EwaldHandlerQuasi2D(), QMCMain::execute(), QMCMain::executeCMCSection(), QMCMain::executeDebugSection(), QMCMain::executeLoop(), AOBasisBuilder< COT >::expandYlm(), EwaldHandler3D::fillFk(), RadialOrbitalSetBuilder< COT >::finalize(), QMCDriverNew::finalize(), SimpleFixedNodeBranch::finalize(), TraceManager::finalize_traces(), KContainer::findApproxMMax(), QMCFixedSampleLinearOptimize::finish(), ForceChiesaPBCAA::ForceChiesaPBCAA(), DensityMatrices1B::generate_density_samples(), DensityMatrices1B::generate_samples(), generateCuspInfo(), OneBodyDensityMatrices::generateDensitySamples(), OneBodyDensityMatrices::generateDensitySamplesWithSpin(), QMCFixedSampleLinearOptimizeBatched::generateSamples(), OneBodyDensityMatrices::generateSamples(), QMCFixedSampleLinearOptimize::generateSamples(), SkAllEstimator::get(), EnergyDensityEstimator::get_particleset(), QMCCostFunction::getConfigurations(), QMCCostFunctionBatched::getConfigurations(), LRCoulombSingleton::getDerivHandler(), LRCoulombSingleton::getHandler(), LinearMethod::getLowestEigenvector(), MPC::init_f_G(), OutputMatrix::init_file(), MPC::init_gvecs(), MPC::init_spline(), MPC::initBreakup(), EwaldHandler3D::initBreakup(), QMCFiniteSize::initBreakup(), LRRPABFeeHandlerTemp< Func, BreakupBasis >::InitBreakup(), LRRPAHandlerTemp< Func, BreakupBasis >::InitBreakup(), LRHandlerTemp< Func, BreakupBasis >::InitBreakup(), CoulombPBCAA::initBreakup(), CoulombPBCAB::initBreakup(), LRHandlerSRCoulomb< Func, BreakupBasis >::InitBreakup(), CSUpdateBase::initCSWalkersForPbyP(), BsplineFunctor< REAL >::initialize(), HybridRepSetReader< SA >::initialize_hybrid_pio_gather(), HybridRepSetReader< SA >::initialize_hybridrep_atomic_centers(), SpaceGrid::initialize_rectilinear(), SplineSetReader< typename SA::SplineBase >::initialize_spline_pio_gather(), QMCHamiltonian::initialize_traces(), TraceManager::initialize_traces(), SpaceGrid::initialize_voronoi(), TimerManager< qmcplusplus::TimerType< CLOCK > >::initializeTimer(), SFNBranch::initParam(), SimpleFixedNodeBranch::initReptile(), RadialJastrowBuilder::initTwoBodyFunctor(), ForceBase::initVarReduction(), SimpleFixedNodeBranch::initWalkerController(), QMCUpdateBase::initWalkersForPbyP(), InitMolecularSystem::initWithVolume(), QMCCostFunctionBase::isEffectiveWeightValid(), kSpaceJastrow::kSpaceJastrow(), LatticeDeviationEstimator::LatticeDeviationEstimator(), LCAOrbitalBuilder::loadBasisSetFromH5(), LCAOrbitalBuilder::loadBasisSetFromXML(), LCAOSpinorBuilder::loadMO(), LCAOrbitalBuilder::loadMO(), RotatedSPOs::log_antisym_matrix(), main(), TraceManager::make_combined_trace(), EnergyDensityEstimator::makeClone(), TraceManager::makeClone(), CloneManager::makeClones(), WalkerLogManager::makeCollector(), BackflowBuilder::makeLongRange_twoBody(), RPAJastrow::makeShortRange(), BackflowBuilder::makeShortRange_twoBody(), QMCDriverNew::measureImbalance(), MomentumDistribution::MomentumDistribution(), EinsplineSetBuilder::OccupyBands(), EinsplineSetBuilder::OccupyBands_ESHDF(), QMCFixedSampleLinearOptimizeBatched::one_shift_run(), TraceManager::open_file(), TraceManager::open_hdf_file(), WalkerLogManager::openFile(), WalkerLogManager::openHDFFile(), operator<<(), ci_configuration::operator==(), PairCorrEstimator::PairCorrEstimator(), parse_pbc_fcc_lattice(), parse_pbc_lattice(), ECPComponentBuilder::parseCasino(), DriftModifierUNR::parseXML(), QMCFixedSampleLinearOptimizeBatched::previous_linear_methods_run(), TimerManager< qmcplusplus::TimerType< CLOCK > >::print(), QMCFixedSampleLinearOptimizeBatched::print_cost_summary(), QMCFixedSampleLinearOptimizeBatched::print_cost_summary_header(), TimerManager< qmcplusplus::TimerType< CLOCK > >::print_flat(), QMCState::print_memory_change(), TimerManager< qmcplusplus::TimerType< CLOCK > >::print_stack(), WaveFunctionTester::printEloc(), QMCCostFunctionBase::printEstimates(), QMCFiniteSize::printSkRawSphAvg(), QMCFiniteSize::printSkSplineSphAvg(), Reptile::printState(), SFNBranch::printStatus(), QMCFixedSampleLinearOptimizeBatched::process(), VMCBatched::process(), DMCBatched::process(), QMCDriver::process(), QMCFixedSampleLinearOptimize::processOptXML(), QMCFixedSampleLinearOptimizeBatched::processOptXML(), QMCMain::processPWH(), HybridEngine::processXML(), DescentEngine::processXML(), LatticeParser::put(), ReferencePoints::put(), VMC::put(), GridExternalPotential::put(), SkPot::put(), InitMolecularSystem::put(), ACForce::put(), EnergyDensityEstimator::put(), CSVMC::put(), RPAJastrow::put(), SkAllEstimator::put(), SpaceGrid::put(), RMCUpdateAllWithDrift::put(), RMCUpdatePbyPWithDrift::put(), NonLocalTOperator::put(), ECPotentialBuilder::put(), RandomNumberControl::put(), ECPComponentBuilder::put(), SPOSetScanner::put(), QMCFixedSampleLinearOptimize::put(), ForceCeperley::put(), WalkerControl::put(), QMCCostFunctionBase::put(), MPC::put(), EstimatorManagerNew::put(), StressPBC::put(), ForceChiesaPBCAA::put(), EstimatorManagerBase::put(), WalkerControlBase::put(), OrbitalImages::put(), PadeFunctor< T >::put(), CountingGaussian::put(), BsplineFunctor< REAL >::put(), Pade2ndOrderFunctor< T >::put(), PadeTwo2ndOrderFunctor< T >::put(), PolynomialFunctor3D::put(), TraceManager::put(), SlaterDetBuilder::putDeterminant(), AOBasisBuilder< COT >::putH5(), eeI_JastrowBuilder::putkids(), LCAOrbitalBuilder::putPBCFromH5(), QMCDriver::putQMCInfo(), MomentumEstimator::putSpecial(), ForwardWalking::putSpecial(), QMCCostFunction::QMCCostFunction(), QMCCostFunctionBatched::QMCCostFunctionBatched(), QMCFiniteSize::QMCFiniteSize(), QMCFixedSampleLinearOptimize::QMCFixedSampleLinearOptimize(), QMCFixedSampleLinearOptimizeBatched::QMCFixedSampleLinearOptimizeBatched(), QMCMain::QMCMain(), HybridEngine::queryStore(), ParticleSetPool::randomize(), ParticleSet::randomizeFromSource(), SimpleFixedNodeBranch::read(), HDFWalkerInput_0_4::read_hdf5(), RandomNumberControl::read_parallel(), HDFWalkerInput_0_4::read_phdf5(), RandomNumberControl::read_rank_0(), PWBasis::readbasis(), SlaterDetBuilder::readCoeffs(), readCuspInfo(), SlaterDetBuilder::readDetList(), SlaterDetBuilder::readDetListH5(), EinsplineSetBuilder::ReadGvectors_ESHDF(), EinsplineSetBuilder::ReadOrbitalInfo(), EinsplineSetBuilder::ReadOrbitalInfo_ESHDF(), ParticleSetPool::readSimulationCellXML(), VMCDriverInput::readXML(), XMLParticleParser::readXML(), CompositeSPOSet::report(), SHOSetBuilder::report(), PairCorrEstimator::report(), SPOInfo::report(), FreeOrbital::report(), StaticStructureFactor::report(), SPOSetInfo::report(), SpinDensity::report(), SPOSetInputInfo::report(), SHOSet::report(), SpinDensityNew::report(), DensityMatrices1B::report(), InputSection::report(), OrbitalImages::report(), TraceRequest::report(), CountingGaussian::reportStatus(), ParticleSet::reset(), TraceManager::reset_buffers(), WalkerLogCollector::resetBuffers(), ParticleSet::resetGroups(), SimulationCell::resetLRBox(), VMC::resetRun(), CSVMC::resetRun(), RMC::resetRun(), SimpleFixedNodeBranch::resetRun(), DMC::resetUpdateEngines(), MCWalkerConfiguration::resetWalkerProperty(), AGPDeterminant::resize(), PWOrbitalSet::resize(), PWRealOrbitalSet::resize(), DensityEstimator::resize(), HybridRepCenterOrbitals< SPLINEBASE::DataType >::resizeStorage(), VMC::run(), RMC::run(), DMC::run(), CSVMC::run(), QMCFixedSampleLinearOptimize::run(), WaveFunctionTester::run(), GradientTest::run(), QMCFixedSampleLinearOptimizeBatched::run(), VMCBatched::run(), DMCBatched::run(), WaveFunctionTester::runBasicTest(), WaveFunctionTester::runCloneTest(), WaveFunctionTester::runDerivCloneTest(), WaveFunctionTester::runDerivNLPPTest(), WaveFunctionTester::runDerivTest(), WaveFunctionTester::runGradSourceTest(), WaveFunctionTester::runNodePlot(), QMCMain::runQMC(), WaveFunctionTester::runRatioTest(), WaveFunctionTester::runRatioTest2(), WaveFunctionTester::runRatioV(), WaveFunctionTester::runZeroVarianceTest(), DescentEngine::sample_finish(), saveCusp(), QMCAppBase::saveXml(), TraceSamples< std::complex< TraceReal > >::screen_writes(), BandInfoGroup::selectBands(), LinearMethod::selectEigenvalue(), EinsplineSetBuilder::set_metadata(), BsplineReader::setCommon(), ECPComponentBuilder::SetQuadratureRule(), QMCDriver::setStatus(), QMCDriverNew::setStatus(), LRBreakup< BreakupBasis >::SetupKVecs(), DescentEngine::setupUpdate(), QMCDriver::setWalkerOffsets(), SHOState::sho_report(), SHOSetBuilder::SHOSetBuilder(), NESpaceGrid< REAL >::someMoreAxisGridStuff(), SpeciesKineticEnergy::SpeciesKineticEnergy(), QMCFixedSampleLinearOptimizeBatched::start(), WalkerLogCollector::startBlock(), TraceManager::startBlock(), WalkerLogManager::startRun(), TraceManager::startRun(), TraceManager::stopBlock(), WalkerLogManager::stopRun(), TraceManager::stopRun(), DescentEngine::storeVectors(), StressPBC::StressPBC(), StructFact::StructFact(), QMCFiniteSize::summary(), SYCLDeviceManager::SYCLDeviceManager(), RandomNumberControl::test(), SpinDensity::test(), TraceBuffer< TraceInt >::test_buffer_collect(), TraceBuffer< TraceInt >::test_buffer_write(), TEST_CASE(), SHOSet::test_derivatives(), DensityMatrices1B::test_derivatives(), test_DiracDeterminant_delayed_update(), test_DiracDeterminant_second(), test_DiracDeterminantBatched_delayed_update(), test_DiracDeterminantBatched_second(), SpinDensity::test_evaluate(), test_J3_polynomial3D(), test_LCAO_DiamondC_2x1x1_cplx(), test_LCAO_DiamondC_2x1x1_real(), test_lcao_spinor(), test_lcao_spinor_excited(), test_lcao_spinor_ion_derivs(), test_LiH_msd(), SHOSet::test_overlap(), DensityMatrices1B::test_overlap(), BackflowTransformation::testDeriv(), DiracDeterminantWithBackflow::testDerivFjj(), SlaterDetWithBackflow::testDerivGL(), DiracDeterminantWithBackflow::testDerivLi(), DiracDeterminantWithBackflow::testGG(), DiracDeterminantWithBackflow::testGGG(), BackflowTransformation::testPbyP(), testTrialWaveFunction_diamondC_2x1x1(), PWBasis::trimforecut(), KContainer::updateKLists(), SFNBranch::updateParamAfterPopControl(), DescentEngine::updateParameters(), TraceSamples< std::complex< TraceReal > >::user_report(), TraceManager::user_report(), ECPotentialBuilder::useSimpleTableFormat(), ECPotentialBuilder::useXmlFormat(), QMCFiniteSize::validateXML(), QMCMain::validateXML(), QMCFixedSampleLinearOptimize::ValidCostFunction(), QMCFixedSampleLinearOptimizeBatched::ValidCostFunction(), WalkerLogManager::WalkerLogManager(), WaveFunctionTester::WaveFunctionTester(), TraceManager::write_buffers(), TraceManager::write_buffers_hdf(), EnergyDensityEstimator::write_Collectables(), EnergyDensityEstimator::write_EDValues(), TraceBuffer< TraceInt >::write_hdf(), EnergyDensityEstimator::write_nonzero_domains(), OrbitalImages::write_orbital_xsf(), TraceRequest::write_selected(), TraceSample< TraceReal >::write_summary(), TraceSamples< std::complex< TraceReal > >::write_summary(), TraceBuffer< TraceInt >::write_summary(), TraceManager::write_summary(), CombinedTraceSample< TraceReal >::write_summary_combined(), WalkerLogManager::writeBuffers(), WalkerLogManager::writeBuffersHDF(), and WalkerLogBuffer< WLog::Real >::writeSummary().
|
inline |
Definition at line 63 of file OutputManager.h.
References InfoStream::getStream(), and infoSummary.
Referenced by HamiltonianFactory::addCoulombPotential(), HamiltonianFactory::addMPCPotential(), HamiltonianFactory::addPseudoPotential(), HamiltonianFactory::build(), JastrowBuilder::buildComponent(), RadialJastrowBuilder::buildComponent(), SlaterDetBuilder::buildComponent(), SPOSetBuilderFactory::buildSPOSetCollection(), WaveFunctionFactory::buildTWF(), DMCFactoryNew::create(), VMCFactoryNew::create(), createBsplineComplex(), createBsplineReal(), RadialJastrowBuilder::createJ1(), RadialJastrowBuilder::createJ2(), SPOSetBuilder::createSPOSet(), LCAOrbitalBuilder::createSPOSetFromXML(), QMCDriverNew::initializeQMC(), QMCAppBase::parse(), LatticeParser::put(), ParticleSetPool::put(), RandomNumberControl::put(), BsplineFunctor< REAL >::put(), ShortRangeCuspFunctor< T >::put(), PolynomialFunctor3D::put(), SlaterDetBuilder::putDeterminant(), QMCMain::QMCMain(), QMCWFOptLinearFactoryNew(), ParticleSetPool::readSimulationCellXML(), DMCDriverInput::readXML(), QMCDriverInput::readXML(), EstimatorManagerInput::readXML(), SimulationCell::resetLRBox(), VMCBatched::run(), DMCBatched::run(), TEST_CASE(), and QMCMain::validateXML().
|
inline |
Definition at line 69 of file OutputManager.h.
References InfoStream::getStream(), and infoLog.
Referenced by CloneManager::acceptRatio(), QMCHamiltonian::addOperator(), HamiltonianFactory::addPseudoPotential(), QMCDriverNew::adjustGlobalWalkerCount(), EinsplineSetBuilder::AnalyzeTwists2(), WalkerControlBase::applyNmaxNmin(), LatticeGaussianProductBuilder::buildComponent(), kSpaceJastrowBuilder::buildComponent(), SlaterDetBuilder::buildComponent(), JastrowBuilder::buildkSpace(), ECPComponentBuilder::buildSemiLocalAndLocal(), TwoBodyJastrow< FT >::checkSanity(), J1OrbitalSoA< FT >::checkSanity(), checkTagStatus(), SlaterDetBuilder::createMSDFast(), PWOrbitalSetBuilder::createPW(), QMCDriverFactory::createQMCDriver(), QMCDriverNew::createRngsStepContexts(), SPOSetBuilder::createSPOSet(), LCAOrbitalBuilder::createSPOSetFromXML(), EinsplineSpinorSetBuilder::createSPOSetFromXML(), EinsplineSetBuilder::createSPOSetFromXML(), CUDADeviceManager::CUDADeviceManager(), WalkerConfigurations::destroyWalkers(), DeviceManager::DeviceManager(), QMCDriverNew::endBlock(), QMCMain::executeLoop(), LCAOrbitalBuilder::LCAOrbitalBuilder(), LCAOrbitalBuilder::loadBasisSetFromXML(), RealSpacePositionsOMPTarget::mw_acceptParticlePos(), OMPDeviceManager::OMPDeviceManager(), LCAOHDFParser::parse(), TimerManager< qmcplusplus::TimerType< CLOCK > >::print_stack(), QMCFixedSampleLinearOptimizeBatched::process(), ParticleSetPool::put(), QMCFixedSampleLinearOptimize::put(), QMCCostFunctionBase::put(), SlaterDetBuilder::putDeterminant(), QMCDriver::putQMCInfo(), RotatedSPOs::readVariationalParameters(), EstimatorManagerInput::readXML(), MCWalkerConfiguration::resize(), kSpaceJastrow::setCoefficients(), MCPopulation::spawnWalker(), SYCLDeviceManager::SYCLDeviceManager(), ECPotentialBuilder::useXmlFormat(), and QMCMain::validateXML().
| void applyCuspCorrection | ( | const Matrix< CuspCorrectionParameters > & | info, |
| ParticleSet & | targetPtcl, | ||
| ParticleSet & | sourcePtcl, | ||
| LCAOrbitalSet & | lcao, | ||
| SoaCuspCorrection & | cusp, | ||
| const std::string & | id | ||
| ) |
Definition at line 568 of file CuspCorrectionConstruction.cpp.
References SoaCuspCorrection::add(), LCAOrbitalSet::C, Matrix< T, Alloc >::cols(), computeRadialPhiBar(), createGlobalTimer(), SPOSet::getOrbitalSetSize(), OutputManagerClass::isDebugActive(), LCAOrbitalSet::isIdentity(), LCAOrbitalSet::isOMPoffload(), LCAOrbitalSet::myBasisSet, outputManager, removeSTypeOrbitals(), Vector< T, Alloc >::resize(), Matrix< T, Alloc >::rows(), Vector< T, Alloc >::size(), OneDimQuinticSpline< Td, Tg, CTd, CTg >::spline(), splitPhiEta(), and timer_level_medium.
Referenced by LCAOrbitalBuilder::createSPOSetFromXML().
| sycl::event qmcplusplus::applyW_stageV_sycl | ( | sycl::queue & | aq, |
| const int *restrict | delay_list_gpu, | ||
| const int | delay_count, | ||
| T *restrict | temp_gpu, | ||
| const int | numorbs, | ||
| const int | ndelay, | ||
| T *restrict | V_gpu, | ||
| const T *restrict | Ainv, | ||
| const std::vector< sycl::event > & | dependencies | ||
| ) |
Definition at line 19 of file sycl_determinant_helper.cpp.
Referenced by DelayedUpdateSYCL< T, T_FP >::updateInvMat().
| sycl::event qmcplusplus::applyW_stageV_sycl | ( | sycl::queue & | aq, |
| const int * | delay_list_gpu, | ||
| const int | delay_count, | ||
| T * | temp_gpu, | ||
| const int | numorbs, | ||
| const int | ndelay, | ||
| T * | V_gpu, | ||
| const T * | Ainv, | ||
| const std::vector< sycl::event > & | dependencies = {} |
||
| ) |
| template sycl::event qmcplusplus::applyW_stageV_sycl | ( | sycl::queue & | aq, |
| const int *restrict | delay_list_gpu, | ||
| const int | delay_count, | ||
| float *restrict | temp_gpu, | ||
| const int | numorbs, | ||
| const int | ndelay, | ||
| float *restrict | V_gpu, | ||
| const float *restrict | Ainv, | ||
| const std::vector< sycl::event > & | dependencies | ||
| ) |
| template sycl::event qmcplusplus::applyW_stageV_sycl | ( | sycl::queue & | aq, |
| const int *restrict | delay_list_gpu, | ||
| const int | delay_count, | ||
| double *restrict | temp_gpu, | ||
| const int | numorbs, | ||
| const int | ndelay, | ||
| double *restrict | V_gpu, | ||
| const double *restrict | Ainv, | ||
| const std::vector< sycl::event > & | dependencies | ||
| ) |
| template sycl::event qmcplusplus::applyW_stageV_sycl | ( | sycl::queue & | aq, |
| const int *restrict | delay_list_gpu, | ||
| const int | delay_count, | ||
| std::complex< float > *restrict | temp_gpu, | ||
| const int | numorbs, | ||
| const int | ndelay, | ||
| std::complex< float > *restrict | V_gpu, | ||
| const std::complex< float > *restrict | Ainv, | ||
| const std::vector< sycl::event > & | dependencies | ||
| ) |
| template sycl::event qmcplusplus::applyW_stageV_sycl | ( | sycl::queue & | aq, |
| const int *restrict | delay_list_gpu, | ||
| const int | delay_count, | ||
| std::complex< double > *restrict | temp_gpu, | ||
| const int | numorbs, | ||
| const int | ndelay, | ||
| std::complex< double > *restrict | V_gpu, | ||
| const std::complex< double > *restrict | Ainv, | ||
| const std::vector< sycl::event > & | dependencies | ||
| ) |
| bool approxEquality | ( | T | val_a, |
| T | val_b, | ||
| std::optional< double > | eps | ||
| ) |
Definition at line 26 of file checkMatrix.hpp.
Referenced by TEST_CASE().
| bool qmcplusplus::approxEquality | ( | const TinyVector< T1, D > & | val_a, |
| const TinyVector< T2, D > & | val_b | ||
| ) |
Definition at line 62 of file test_ReferencePoints.cpp.
| template bool approxEquality< double > | ( | double | val_a, |
| double | val_b, | ||
| std::optional< double > | eps | ||
| ) |
| template bool approxEquality< float > | ( | float | val_a, |
| float | val_b, | ||
| std::optional< double > | eps | ||
| ) |
| template bool approxEquality< std::complex< double > > | ( | std::complex< double > | val_a, |
| std::complex< double > | val_b, | ||
| std::optional< double > | eps | ||
| ) |
| template bool approxEquality< std::complex< float > > | ( | std::complex< float > | val_a, |
| std::complex< float > | val_b, | ||
| std::optional< double > | eps | ||
| ) |
|
inline |
Definition at line 40 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by FnArcSin::operator()(), and RPABFeeBreakup< T >::Xk().
|
inline |
Definition at line 1387 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Definition at line 736 of file OhmmsMatrixOperators.h.
References evaluate().
Referenced by Matrix< ST, qmcplusplus::Mallocator< ST > >::copy(), Matrix< ST, qmcplusplus::Mallocator< ST > >::Matrix(), ParticleAttrib< qmcplusplus::TinyVector >::operator=(), Vector< T, std::allocator< T > >::operator=(), Matrix< ST, qmcplusplus::Mallocator< ST > >::operator=(), and XMLNodeString::XMLNodeString().
|
inline |
Definition at line 769 of file ParticleAttrib.cpp.
References evaluate().
Definition at line 2225 of file OhmmsVectorOperators.h.
References evaluate().
|
inline |
Definition at line 203 of file DriftOperators.h.
References qmcplusplus::Units::time::s.
Referenced by VMCUpdateAll::advanceWalker(), CSVMCUpdateAllWithDrift::advanceWalker(), RMCUpdateAllWithDrift::advanceWalkersRMC(), and RMCUpdateAllWithDrift::advanceWalkersVMC().
|
inline |
|
inline |
Definition at line 219 of file DriftOperators.h.
References getScaledDrift().
|
inline |
Definition at line 33 of file RandomSeqGenerator.h.
References cos(), log(), n, sin(), and sqrt().
Referenced by OneBodyDensityMatrices::diffuse(), OneBodyDensityMatrices::diffuseSpin(), DensityMatrices1B::diffusion(), makeGaussRandom(), makeGaussRandomWithEngine(), and TEST_CASE().
|
inline |
Definition at line 64 of file RandomSeqGenerator.h.
References n.
Referenced by makeUniformRandom(), and randomize().
|
inline |
Definition at line 48 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by CartesianTensor< T, Point_t, Tensor_t, GGG_t >::CartesianTensor(), SphericalTensor< T, Point_t, Tensor_t, GGG_t >::evaluate(), SoaSphericalTensor< ST >::evaluate_bare(), SphericalTensor< T, Point_t, Tensor_t, GGG_t >::evaluateAll(), SphericalTensor< T, Point_t, Tensor_t, GGG_t >::evaluateTest(), SphericalTensor< T, Point_t, Tensor_t, GGG_t >::evaluateWithHessian(), GaussianCombo< T >::GaussianCombo(), FnArcTan::operator()(), Quadrature3D< T >::Quadrature3D(), SoaCartesianTensor< T >::SoaCartesianTensor(), SoaSphericalTensor< ST >::SoaSphericalTensor(), and SphericalTensor< T, Point_t, Tensor_t, GGG_t >::SphericalTensor().
|
inline |
Definition at line 1395 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 338 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by NESpaceGrid< REAL >::accumulate(), cart2sph(), SpaceGrid::check_grid(), NESpaceGrid< REAL >::check_grid(), derivYlmSpherical(), SpaceGrid::evaluate(), fix_phase_rotate(), FnArcTan2::operator()(), and Ylm().
|
inline |
Definition at line 588 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 838 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1062 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1263 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1685 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1909 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 2110 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 78 of file CommUtilities.h.
Referenced by EinsplineSetBuilder::bcastSortBands(), RandomNumberControl::make_seeds(), RandomNumberControl::put(), HDFWalkerInput_0_4::read_hdf5_scatter(), HDFWalkerInput_0_4::read_phdf5(), and RandomNumberControl::read_rank_0().
| void qmcplusplus::bessel_steed_array_cpu | ( | const int | lmax, |
| const T | x, | ||
| T * | jl | ||
| ) |
Compute spherical bessel function from 0 to lmax.
Using Steed/Barnett algorithm from Comp. Phys. Comm. 21, 297 (1981)
Definition at line 25 of file Bessel.h.
References B(), BLAS::cone, and qmcplusplus::simd::inv().
Referenced by Gvectors< ST, LT >::calc_jlm_G(), and TEST_CASE().
| qmcplusplus::BOOSTSUB_H5_DATATYPE | ( | char | , |
| H5T_NATIVE_CHAR | |||
| ) |
| qmcplusplus::BOOSTSUB_H5_DATATYPE | ( | short | , |
| H5T_NATIVE_SHORT | |||
| ) |
| qmcplusplus::BOOSTSUB_H5_DATATYPE | ( | int | , |
| H5T_NATIVE_INT | |||
| ) |
| qmcplusplus::BOOSTSUB_H5_DATATYPE | ( | long | , |
| H5T_NATIVE_LONG | |||
| ) |
| qmcplusplus::BOOSTSUB_H5_DATATYPE | ( | long | long, |
| H5T_NATIVE_LLONG | |||
| ) |
| qmcplusplus::BOOSTSUB_H5_DATATYPE | ( | unsigned | char, |
| H5T_NATIVE_UCHAR | |||
| ) |
| qmcplusplus::BOOSTSUB_H5_DATATYPE | ( | unsigned | short, |
| H5T_NATIVE_USHORT | |||
| ) |
| qmcplusplus::BOOSTSUB_H5_DATATYPE | ( | unsigned | int, |
| H5T_NATIVE_UINT | |||
| ) |
| qmcplusplus::BOOSTSUB_H5_DATATYPE | ( | unsigned | long, |
| H5T_NATIVE_ULONG | |||
| ) |
| qmcplusplus::BOOSTSUB_H5_DATATYPE | ( | unsigned long | long, |
| H5T_NATIVE_ULLONG | |||
| ) |
| qmcplusplus::BOOSTSUB_H5_DATATYPE | ( | float | , |
| H5T_NATIVE_FLOAT | |||
| ) |
| qmcplusplus::BOOSTSUB_H5_DATATYPE | ( | double | , |
| H5T_NATIVE_DOUBLE | |||
| ) |
| void broadcastCuspInfo | ( | CuspCorrectionParameters & | param, |
| Communicate & | Comm, | ||
| int | root | ||
| ) |
Broadcast cusp correction parameters.
Definition at line 198 of file CuspCorrectionConstruction.cpp.
References CuspCorrectionParameters::alpha, CuspCorrectionParameters::C, CuspCorrectionParameters::Rc, CuspCorrectionParameters::redo, and CuspCorrectionParameters::sg.
Referenced by LCAOrbitalBuilder::createSPOSetFromXML(), generateCuspInfo(), and TEST_CASE().
|
inline |
Definition at line 282 of file SmallMatrixDetCalculator.h.
References evaluate(), and n.
Referenced by MultiDiracDeterminant::buildTableMatrix_calculateRatios_impl(), and TestSmallMatrixDetCalculator< T >::customized_evaluate().
|
inline |
Definition at line 74 of file CommUtilities.h.
| CUDATypeMap<T> qmcplusplus::castCUDAType | ( | T | var | ) |
Definition at line 53 of file CUDATypeMapping.hpp.
Referenced by qmcplusplus::cuBLAS::getrf_batched(), and qmcplusplus::cuBLAS::getri_batched().
| hipblasTypeMap<T> qmcplusplus::casthipblasType | ( | T | var | ) |
Definition at line 48 of file hipblasTypeMapping.hpp.
|
inline |
Definition at line 56 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by ECPComponentBuilder::buildL2(), ECPComponentBuilder::buildSO(), SpinDensityInput::calculateDerivedParameters(), MagnetizationDensityInput::calculateDerivedParameters(), SHOSetBuilder::createSPOSet(), FreeOrbitalBuilder::createSPOSetFromXML(), ECPComponentBuilder::doBreakUp(), OptimalGrid::Init(), HybridRepSetReader< SA >::initialize_hybridrep_atomic_centers(), FnCeil::operator()(), VMC::put(), CSVMC::put(), SpinDensity::put(), PairCorrEstimator::put(), EinsplineSetBuilder::ReadGvectors_ESHDF(), VMC::resetRun(), and SHOSetBuilder::update_basis_states().
|
inline |
Definition at line 1403 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
| qmcplusplus::CHECK | ( | oh. | lower_bound = =0 | ) |
| CHECK | ( | embt.em. | getNumEstimators() = =0 | ) |
| CHECK | ( | emi. | get_estimator_inputs).size( = =2 | ) |
| CHECK | ( | emn. | getNumEstimators() = =2 | ) |
| qmcplusplus::CHECK | ( | emnta. | getMainEstimator).getName( = ="LocalEnergyEstimator" | ) |
| CHECK | ( | emi2. | get_estimator_inputs).size( = =2 | ) |
| CHECK | ( | emn2. | getNumEstimators() = =2 | ) |
| qmcplusplus::CHECK | ( | emnta2. | getMainEstimator).getName( = ="RMCLocalEnergyEstimator" | ) |
| qmcplusplus::CHECK | ( | log_values | [0] = =ComplexApprox(std::complex< double >{ 5.603777579195571, -6.1586603331188225 }) | ) |
Referenced by check_force_copy(), check_matrix(), OneBodyDensityMatricesTests< T >::checkData(), complex_test_case(), doSOECPotentialTest(), double_test_case(), fill_from_text(), MinTest::find_min(), getTestCaseForWeights(), if(), mpiTestFunctionWrapped(), TestFillBufferRngReal< T >::operator()(), TestFillVecRngReal< T >::operator()(), TestGetVecRngReal< T >::operator()(), TestFillBufferRngComplex< T >::operator()(), TestFillVecRngComplex< T >::operator()(), TestGetVecRngComplex< T >::operator()(), TEMPLATE_TEST_CASE(), test_C_diamond(), test_cartesian_ao(), TEST_CASE(), optimize::TEST_CASE(), qmcplusplus::testing::TEST_CASE(), TEST_CASE(), test_CoulombPBCAA_3p(), test_diamond_2x1x1_xml_input(), test_dirac_ao(), test_DiracDeterminant_delayed_update(), test_DiracDeterminant_first(), test_DiracDeterminant_second(), test_DiracDeterminantBatched_delayed_update(), test_DiracDeterminantBatched_first(), test_DiracDeterminantBatched_second(), test_distance_fcc_pbc_z_batched_APIs(), test_distance_pbc_z_batched_APIs(), test_distance_pbc_z_batched_APIs_ee_NEED_TEMP_DATA_ON_HOST(), test_e2iphi(), test_EtOH_mw(), test_gemm(), test_gemv(), test_gemv_batched(), test_ger(), test_ger_batched(), test_HCN(), test_hcpBe_rotation(), test_He(), test_He_mw(), test_He_sto3g_xml_input(), test_inverse(), test_isnan(), test_J1_spline(), test_J3_polynomial3D(), test_LCAO_DiamondC_2x1x1_cplx(), test_LCAO_DiamondC_2x1x1_real(), test_lcao_spinor(), test_lcao_spinor_excited(), test_lcao_spinor_ion_derivs(), test_LiH_msd(), test_Ne(), test_one_gemm(), test_one_gemv(), test_one_ger(), test_real_accumulator(), test_real_accumulator_basic(), test_real_accumulator_weights(), test_spline_bounds(), test_tiny_vector(), test_tiny_vector_size_two(), QMCDriverNewTestWrapper::testAdjustGlobalWalkerCount(), SpinDensityNewTests::testCopyConstructor(), MomentumDistributionTests::testCopyConstructor(), MagnetizationDensityTests::testCopyConstructor(), OneBodyDensityMatricesTests< T >::testCopyConstructor(), MagnetizationDensityTests::testData(), QMCDriverNewTestWrapper::testDetermineStepsPerBlock(), testDualAllocator(), OneBodyDensityMatricesTests< T >::testGenerateSamples(), OneBodyDensityMatricesTests< T >::testGenerateSamplesForSpinor(), MagnetizationDensityTests::testGrids(), MagnetizationDensityTests::testIntegrationFunctions(), EstimatorManagerNewTest::testReplaceMainEstimator(), and testTrialWaveFunction_diamondC_2x1x1().
| qmcplusplus::CHECK | ( | log_values | [1] = =ComplexApprox(std::complex< double >{ 5.531331998282581, -8.805487075984523 }) | ) |
| void qmcplusplus::check_force_copy | ( | ForceChiesaPBCAA & | force, |
| ForceChiesaPBCAA & | force2 | ||
| ) |
Definition at line 86 of file test_force.cpp.
References ForceChiesaPBCAA::c, CHECK(), Matrix< T, Alloc >::cols(), ForceBase::getAddIonIon(), ForceChiesaPBCAA::getDistanceTableAAID(), ForceBase::getForcesIonIon(), ForceChiesaPBCAA::h, ForceChiesaPBCAA::m_exp, ForceChiesaPBCAA::N_basis, ForceChiesaPBCAA::NptclA, ForceChiesaPBCAA::NptclB, ForceChiesaPBCAA::NumSpeciesA, ForceChiesaPBCAA::NumSpeciesB, ForceChiesaPBCAA::Qat, ForceChiesaPBCAA::Qspec, ForceChiesaPBCAA::Rcut, REQUIRE(), Matrix< T, Alloc >::rows(), ForceChiesaPBCAA::Sinv, Matrix< T, Alloc >::size(), Vector< T, Alloc >::size(), ForceChiesaPBCAA::Zat, and ForceChiesaPBCAA::Zspec.
Referenced by TEST_CASE().
Definition at line 39 of file test_DiracDeterminant.cpp.
References CHECK(), Matrix< T, Alloc >::cols(), REQUIRE(), Matrix< T, Alloc >::rows(), and Matrix< T, Alloc >::size().
Referenced by test_DiracDeterminant_delayed_update(), test_DiracDeterminant_first(), and test_DiracDeterminant_second().
| qmcplusplus::checkArray | ( | real_pivot | , |
| pivots | , | ||
| 8 | |||
| ) |
| CHECKED_ELSE | ( | check_matrix_result. | result | ) |
Definition at line 455 of file test_cuBLAS_LU.cpp.
References check_matrix_result.
Referenced by TEMPLATE_TEST_CASE(), TEST_CASE(), test_DiracDeterminantBatched_delayed_update(), test_DiracDeterminantBatched_first(), test_DiracDeterminantBatched_second(), test_inverse(), and testDualAllocator().
| CheckMatrixResult qmcplusplus::checkMatrix | ( | M1 & | a_mat, |
| M2 & | b_mat, | ||
| const bool | check_all = false, |
||
| std::optional< const double > | eps = std::nullopt |
||
| ) |
This function checks equality a_mat and b_mat elements M1, M2 need to have their element type declared M1::value_type and have an operator(i,j) accessor.
I leave the c++14 template meta programming to insure this as an exercise for the reader. Or just enjoy the compiler error.
| [in] | a_mat | - reference matrix, if padded must be identical to b_mat, can be a smaller than b_mat in which case it is compared to upper left block of b_mat. |
| [in] | b_mat | - the matrix to check |
| [in] | check_all | - if true continue to check matrix elements after failure |
| [in] | eps | - add a tolerance for Catch Approx checks. Default to same as in Approx. |
Definition at line 63 of file checkMatrix.hpp.
Referenced by TEMPLATE_TEST_CASE(), TEST_CASE(), test_DiracDeterminantBatched_delayed_update(), test_DiracDeterminantBatched_first(), test_DiracDeterminantBatched_second(), test_inverse(), and testDualAllocator().
| qmcplusplus::checkRs | ( | mc_coords_rs. | positions | ) |
| qmcplusplus::checkRs | ( | mc_coords_rsspins. | positions | ) |
|
inline |
free function to check dimension
| grp | group id |
| aname | name of the dataspace |
| rank | rank of the multi-dimensional array |
| dims[rank] | size for each direction, return the actual size on file |
Definition at line 98 of file hdf_wrapper_functions.h.
References dims, is_same(), and rank.
| bool qmcplusplus::checkVec | ( | T & | vec, |
| T | expected_vec | ||
| ) |
Definition at line 53 of file test_SpaceGridInput.cpp.
Referenced by TEST_CASE().
Definition at line 950 of file TensorOps.h.
References sqrt().
Referenced by DMCUpdatePbyPL2::advanceWalker().
| hFile qmcplusplus::close | ( | ) |
| void qmcplusplus::complex_test_case | ( | ) |
Definition at line 60 of file test_attrib_ops.cpp.
References OTCDot< T1, T2, D >::apply(), OTCDot_CC< T1, T2, D >::apply(), CHECK(), Dot(), Dot_CC(), REQUIRE(), Vector< T, Alloc >::resize(), and Vector< T, Alloc >::size().
| void compute_batch_parameters | ( | int | sample_size, |
| int | batch_size, | ||
| int & | num_batches, | ||
| int & | final_batch_size | ||
| ) |
Compute number of batches and final batch size given the number of samples and a batch size.
| [in] | sample_size | number of samples to process. |
| [in] | batch_size | process samples in batch_size at a time (typically the number of walkers in a crowd). |
| [out] | num_batches | number of batches to use. |
| [out] | final_batch_size | the last batch size. May be smaller than batch_size if the number of samples is not a multiple of the batch size. |
There may be cases where the batch size is zero. One cause is when the number of walkers per rank is less than the number of crowds.
Definition at line 207 of file QMCCostFunctionBatched.cpp.
References batch_size.
Referenced by QMCCostFunctionBatched::checkConfigurations(), QMCCostFunctionBatched::correlatedSampling(), and TEST_CASE().
|
inline |
Compute the norm of an orbital.
| cG | the plane wave coefficients |
Definition at line 220 of file einspline_helper.hpp.
References real(), and sqrt().
Referenced by SplineSetReader< typename SA::SplineBase >::readOneOrbitalCoefs().
|
inline |
Compute the phase factor for rotation.
The algorithm aims at balanced real and imaginary parts.
| in | the real space orbital value on a 3D grid |
| twist | k-point in reduced coordinates |
| phase_r | output real part of the phase |
| phase_i | output imaginary part of the phase |
Definition at line 236 of file einspline_helper.hpp.
References TinyVector< T, D >::data(), qmcplusplus::Units::time::s, sincos(), and sqrt().
Referenced by fix_phase_rotate_c2c().
|
inline |
Definition at line 100 of file DiracMatrix.h.
Referenced by DiracMatrixComputeOMPTarget< VALUE_FP >::computeInvertAndLog(), DiracMatrix< VALUE_FP >::computeInvertAndLog(), cuSolverInverter< T_FP >::invert_transpose(), and rocSolverInverter< T_FP >::invert_transpose().
| std::complex<T> qmcplusplus::computeLogDet_sycl | ( | sycl::queue & | aq, |
| int | n, | ||
| int | lda, | ||
| const TMAT * | a, | ||
| const INDEX * | pivot, | ||
| const std::vector< sycl::event > & | dependencies = {} |
||
| ) |
| std::complex<T> qmcplusplus::computeLogDet_sycl | ( | sycl::queue & | aq, |
| int | n, | ||
| int | lda, | ||
| const TMAT *restrict | a, | ||
| const INDEX *restrict | pivot, | ||
| const std::vector< sycl::event > & | dependencies | ||
| ) |
Definition at line 92 of file sycl_determinant_helper.cpp.
References qmcplusplus::Units::distance::A, lda, log(), and n.
| template std::complex<double> qmcplusplus::computeLogDet_sycl | ( | sycl::queue & | aq, |
| int | n, | ||
| int | lda, | ||
| const double *restrict | a, | ||
| const std::int64_t *restrict | pivot, | ||
| const std::vector< sycl::event > & | dependencies | ||
| ) |
| template std::complex<double> qmcplusplus::computeLogDet_sycl | ( | sycl::queue & | aq, |
| int | n, | ||
| int | lda, | ||
| const std::complex< double > *restrict | a, | ||
| const std::int64_t *restrict | pivot, | ||
| const std::vector< sycl::event > & | dependencies | ||
| ) |
| void computeRadialPhiBar | ( | ParticleSet * | targetP, |
| ParticleSet * | sourceP, | ||
| int | curOrb_, | ||
| int | curCenter_, | ||
| SPOSet * | Phi, | ||
| Vector< QMCTraits::RealType > & | xgrid, | ||
| Vector< QMCTraits::RealType > & | rad_orb, | ||
| const CuspCorrectionParameters & | data | ||
| ) |
Compute the radial part of the corrected wavefunction.
Definition at line 280 of file CuspCorrectionConstruction.cpp.
References OneMolecularOrbital::changeOrbital(), phiBar(), and Vector< T, Alloc >::size().
Referenced by applyCuspCorrection(), and TEST_CASE().
|
inline |
Workaround to allow conj on scalar to return real instead of complex.
Definition at line 96 of file complex_help.hpp.
Referenced by calcAngInt(), OneBodyDensityMatrices::calcDensity(), OneBodyDensityMatrices::calcDensityDrift(), OneBodyDensityMatrices::calcDensityDriftWithSpin(), OneBodyDensityMatrices::calcDensityWithSpin(), SOECPComponent::calculateProjector(), Quadrature3D< T >::CheckQuadratureRule(), RotatedSPOs::constructAntiSymmetricMatrix(), DensityMatrices1B::density_drift(), DensityMatrices1B::density_only(), derivYlmSpherical(), kSpaceJastrow::Equivalent(), kSpaceJastrow::evalGrad(), DensityMatrices1B::evaluate_check(), DensityMatrices1B::evaluate_loop(), kSpaceJastrow::evaluateDerivatives(), kSpaceJastrow::evaluateLog(), SOECPComponent::evaluateOneExactSpinIntegration(), kSpaceJastrow::evaluateRatiosAlltoOne(), evaluateValueAndDerivative(), SOECPComponent::evaluateValueAndDerivatives(), QMCCostFunction::fillOverlapHamiltonianMatrices(), QMCCostFunctionBatched::fillOverlapHamiltonianMatrices(), DensityMatrices1B::generate_particle_basis(), DensityMatrices1B::generate_sample_ratios(), OneBodyDensityMatrices::generateParticleBasis(), OneBodyDensityMatrices::generateSampleRatios(), DensityMatrices1B::integrate(), DensityMatrices1B::normalize(), OneBodyDensityMatrices::normalizeBasis(), kSpaceJastrow::operator()(), kSpaceJastrow::ratio(), kSpaceJastrow::ratioGrad(), and DensityMatrices1B::test_overlap().
|
inline |
Definition at line 97 of file complex_help.hpp.
|
inline |
Definition at line 98 of file complex_help.hpp.
References conj().
|
inline |
Definition at line 99 of file complex_help.hpp.
Referenced by conj().
| void qmcplusplus::convert | ( | const PL & | lat, |
| const PV & | pin, | ||
| PV & | pout | ||
| ) |
Definition at line 24 of file ParticleUtility.h.
Referenced by RadialOrbitalSetBuilder< COT >::finalize(), SPOSetInputInfo::put(), DiracMatrixComputeOMPTarget< VALUE_FP >::reset(), and OrbitalImages::write_orbital_xsf().
| void qmcplusplus::convert2Cart | ( | const PL & | lat, |
| PV & | pin | ||
| ) |
Definition at line 49 of file ParticleUtility.h.
| void qmcplusplus::convert2Unit | ( | const PL & | lat, |
| PV & | pin | ||
| ) |
Definition at line 62 of file ParticleUtility.h.
|
static |
Definition at line 103 of file template_types.hpp.
| FakeChronoClock::duration convert_to_ns | ( | T | in | ) |
Definition at line 27 of file test_runtime_manager.cpp.
Referenced by TEST_CASE().
| double qmcplusplus::convert_to_s | ( | T | in | ) |
Definition at line 46 of file test_timer.cpp.
Referenced by TEST_CASE().
|
static |
Definition at line 92 of file template_types.hpp.
|
static |
Definition at line 140 of file template_types.hpp.
Referenced by QMCCostFunctionBatched::checkConfigurations(), and TEST_CASE().
|
static |
Definition at line 155 of file template_types.hpp.
|
inline |
extract the contents of a string to a vector of something. separator is white spaces.
Definition at line 152 of file string_utils.h.
References qmcplusplus::Units::time::s.
|
inline |
generic conversion from type T1 to type T2 using implicit conversion
Definition at line 32 of file ConvertToReal.h.
Referenced by TWFFastDerivWrapper::calcJastrowRatioGrad(), SpaceWarpTransformation::computeSWT(), convertToReal(), PWRealOrbitalSet::evaluate_notranspose(), TrialWaveFunction::evaluateDeltaLogSetup(), DiracDeterminantWithBackflow::evaluateDerivatives(), kSpaceJastrow::evaluateDerivatives(), QMCHamiltonian::evaluateIonDerivs(), QMCHamiltonian::evaluateIonDerivsDeterministic(), QMCHamiltonian::evaluateIonDerivsDeterministicFast(), TWFFastDerivWrapper::evaluateJastrowRatio(), NonLocalECPComponent::evaluateOneWithForces(), BareKineticEnergy::evaluateWithIonDerivs(), DriftModifierUNR::getDrift(), getScaledDrift(), getScaledDriftL2(), getUnscaledDrift(), setScaledDrift(), setScaledDriftPbyPandNodeCorr(), and TEST_CASE().
|
inline |
specialization of conversion from complex to real
Definition at line 40 of file ConvertToReal.h.
|
inline |
Definition at line 49 of file ConvertToReal.h.
References convertToReal().
specialization for D tensory
Definition at line 57 of file ConvertToReal.h.
References convertToReal().
|
inline |
generic function to convert arrays
| in | starting address of type T1 |
| out | starting address of type T2 |
| n | size of in/out |
Definition at line 69 of file ConvertToReal.h.
References convertToReal(), and n.
specialization for a vector
Definition at line 77 of file ConvertToReal.h.
References convertToReal(), Vector< T, Alloc >::data(), and Vector< T, Alloc >::size().
specialization for a vector
Definition at line 84 of file ConvertToReal.h.
References convertToReal(), Matrix< T, Alloc >::data(), and Matrix< T, Alloc >::size().
|
static |
Definition at line 114 of file template_types.hpp.
|
static |
convert a vector of std::unique_ptrs<T> to a refvector<T>
Definition at line 66 of file template_types.hpp.
Referenced by QMCCostFunctionBatched::correlatedSampling(), EstimatorManagerCrowd::get_operator_estimators(), EstimatorManagerCrowd::get_scalar_estimators(), SetupSFNBranch::operator()(), EstimatorManagerNew::reduceOperatorEstimators(), TEST_CASE(), and RandomNumberControl::write().
|
static |
convert a vector of std::unique_ptrs<T> to a refvector<T2> the static assert prevents ambiguity between this function and the above when T is a derived type of T2.
Definition at line 80 of file template_types.hpp.
|
static |
Definition at line 125 of file template_types.hpp.
Referenced by CostFunctionCrowdData::get_h0_list(), CostFunctionCrowdData::get_h_list(), CostFunctionCrowdData::get_p_list(), and CostFunctionCrowdData::get_wf_list().
|
inline |
evaluate log(psi) as log(|psi|) and phase
| psi | real/complex value |
The return value is always complex regardless of the type of psi. The sign of of a real psi value is represented in the complex part of the return value. The phase of std::log(complex) is in range [-pi, pi] defined in C++
Definition at line 85 of file OrbitalSetTraits.h.
References log().
Referenced by AGPDeterminant::acceptMove(), DiracDeterminantWithBackflow::acceptMove(), DiracDeterminant< DU_TYPE >::acceptMove(), MultiSlaterDetTableMethod::acceptMove(), MultiDiracDeterminant::acceptMove(), DiracDeterminantBatched< PL, VT, FPVT >::acceptMove(), TrialWaveFunction::calcRatioGrad(), TrialWaveFunction::calcRatioGradWithSpin(), MultiSlaterDetTableMethod::evaluate_vgl_impl(), LinearOrbital::evaluateLog(), MultiSlaterDetTableMethod::mw_accept_rejectMove(), and DiracDeterminantBatched< PL, VT, FPVT >::mw_accept_rejectMove().
|
inline |
Definition at line 91 of file OrbitalSetTraits.h.
References log().
|
inline |
|
inline |
Definition at line 214 of file ParticleAttribOps.h.
|
inline |
Definition at line 101 of file complex_help.hpp.
Referenced by RotatedSPOs::exponentiate_antisym_matrix().
|
inline |
Definition at line 102 of file complex_help.hpp.
|
inline |
Definition at line 103 of file complex_help.hpp.
|
inline |
Definition at line 104 of file complex_help.hpp.
| void qmcplusplus::copyGridUnrotatedForTest | ( | NonLocalECPComponent & | nlpp | ) |
Definition at line 153 of file test_ecp.cpp.
References NonLocalECPComponent::rrotsgrid_m, and NonLocalECPComponent::sgridxyz_m.
Referenced by TEST_CASE().
| void qmcplusplus::copyGridUnrotatedForTest | ( | SOECPComponent & | sopp | ) |
Definition at line 154 of file test_ecp.cpp.
References SOECPComponent::rrotsgrid_m_, and SOECPComponent::sgridxyz_m_.
|
inline |
Definition at line 64 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by assignGaussRand(), QMCFiniteSize::build_spherical_grid(), CubicFormula(), LPQHI_BasisClass::dEminus_dk(), LPQHI_BasisClass::dEplus_dk(), derivYlmSpherical(), LPQHIBasis::Eminus(), LPQHISRCoulombBasis::Eminus(), LPQHI_BasisClass::Eminus(), LPQHISRCoulombBasis::Eminus_dG(), LPQHIBasis::Eplus(), LPQHISRCoulombBasis::Eplus(), LPQHI_BasisClass::Eplus(), LPQHISRCoulombBasis::Eplus_dG(), SpinorSet::evaluate_notranspose(), SpinorSet::evaluate_notranspose_spin(), SpinorSet::evaluate_spin(), SpinorSet::evaluateDetSpinorRatios(), SpinorSet::evaluateGradSource(), SpinorSet::evaluateValue(), SpinorSet::evaluateVGL(), SpinorSet::evaluateVGL_spin(), CoulombFunctor< T >::Fk(), generateRotationMatrix(), MagnetizationDensity::generateSpinIntegrand(), MPC::init_f_G(), SpaceGrid::initialize_rectilinear(), ChannelPotential::jl(), kSpaceJastrow::kSpaceJastrow(), SOECPComponent::matrixElementDecomposed(), SpinorSet::mw_evaluate_notranspose(), SpinorSet::mw_evaluateVGLandDetRatioGradsWithSpin(), SpinorSet::mw_evaluateVGLWithSpin(), FnCos::operator()(), KspaceEwaldTerm::operator()(), SFNBranch::phaseChanged(), SimpleFixedNodeBranch::phaseChanged(), Quadrature3D< T >::Quadrature3D(), WaveFunctionTester::runBasicTest(), WaveFunctionTester::runZeroVarianceTest(), sincos(), sMatrixElement(), SOECPComponent::sMatrixElements(), NESpaceGrid< REAL >::someMoreAxisGridStuff(), sph2cart(), kSpaceJastrow::StructureFactor(), TEST_CASE(), test_e2iphi(), EslerCoulomb3D::Xk(), CoulombFunctor< T >::Xk(), YukawaBreakup< T >::Xk(), DerivYukawaBreakup< T >::Xk(), Ylm(), and Ylm().
|
inline |
Definition at line 1411 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 72 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by FnHypCos::operator()().
|
inline |
Definition at line 1419 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 145 of file ParticleAttribOps.h.
References OTCDot< T1, T2, D >::cplx_apply().
Referenced by BareKineticEnergy::evaluate().
|
inline |
Definition at line 192 of file ParticleAttribOps.h.
Referenced by BareKineticEnergy::evaluate().
| hFile qmcplusplus::create | ( | filename | ) |
Referenced by OneSplineOrbData::create(), and HybridRepSetReader< SA >::create_atomic_centers_Gspace().
| void qmcplusplus::create_C_pbc_particlesets | ( | ParticleSet & | elec, |
| ParticleSet & | ions | ||
| ) |
Definition at line 95 of file test_ion_derivs.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), ParticleSet::create(), ParticleSet::createSK(), qmcplusplus::Units::charge::e, ParticleSet::getSpeciesSet(), ParticleSet::R, ParticleSet::resetGroups(), ParticleSet::setName(), and ParticleSet::update().
| QMCHamiltonian& qmcplusplus::create_CN_Hamiltonian | ( | HamiltonianFactory & | hf | ) |
Definition at line 141 of file test_ion_derivs.cpp.
References doc, HamiltonianFactory::getH(), Libxml2Document::getRoot(), okay, Libxml2Document::parseFromString(), HamiltonianFactory::put(), and REQUIRE().
Referenced by TEST_CASE().
| void qmcplusplus::create_CN_particlesets | ( | ParticleSet & | elec, |
| ParticleSet & | ions | ||
| ) |
Definition at line 30 of file test_ion_derivs.cpp.
References ParticleSet::addTable(), doc, Libxml2Document::getRoot(), okay, Libxml2Document::parseFromString(), XMLParticleParser::readXML(), REQUIRE(), and ParticleSet::update().
Referenced by TEST_CASE().
|
static |
Definition at line 47 of file TrialWaveFunction.cpp.
References suffixes.
| UPtrVector<ParticleSet::ParticleGradient> qmcplusplus::create_particle_gradient | ( | int | nelec, |
| int | nentry | ||
| ) |
Definition at line 199 of file test_TrialWaveFunction_He.cpp.
Referenced by TEST_CASE().
| UPtrVector<ParticleSet::ParticleLaplacian> qmcplusplus::create_particle_laplacian | ( | int | nelec, |
| int | nentry | ||
| ) |
Definition at line 208 of file test_TrialWaveFunction_He.cpp.
Referenced by TEST_CASE().
| std::unique_ptr<BsplineReader> qmcplusplus::createBsplineComplex | ( | EinsplineSetBuilder * | e, |
| bool | hybrid_rep, | ||
| const std::string & | useGPU | ||
| ) |
create a reader which handles complex (double size real) splines, C2R or C2C case spline storage and computation precision is ST
Definition at line 24 of file createBsplineComplex.cpp.
References app_summary(), qmcplusplus::Units::charge::e, OHMMS_PRECISION, OMPTARGET, and PlatformSelector< KIND >::selectPlatform().
Referenced by EinsplineSpinorSetBuilder::createSPOSetFromXML(), and EinsplineSetBuilder::createSPOSetFromXML().
| std::unique_ptr< BsplineReader > createBsplineComplex | ( | EinsplineSetBuilder * | e, |
| bool | use_single, | ||
| bool | hybrid_rep, | ||
| const std::string & | useGPU | ||
| ) |
create a reader which handles complex (double size real) splines, C2R or C2C case spline storage and computation precision is double
Definition at line 83 of file createBsplineComplex.cpp.
References qmcplusplus::Units::charge::e.
| std::unique_ptr<BsplineReader> qmcplusplus::createBsplineReal | ( | EinsplineSetBuilder * | e, |
| bool | hybrid_rep, | ||
| const std::string & | useGPU | ||
| ) |
create a reader which handles real splines, R2R case spline storage and computation precision is ST
Definition at line 24 of file createBsplineReal.cpp.
References app_summary(), qmcplusplus::Units::charge::e, OMPTARGET, and PlatformSelector< KIND >::selectPlatform().
| std::unique_ptr< BsplineReader > createBsplineReal | ( | EinsplineSetBuilder * | e, |
| bool | use_single, | ||
| bool | hybrid_rep, | ||
| const std::string & | useGPU | ||
| ) |
create a reader which handles real splines, R2R case spline storage and computation precision is double
Definition at line 44 of file createBsplineReal.cpp.
References qmcplusplus::Units::charge::e.
Referenced by EinsplineSpinorSetBuilder::createSPOSetFromXML(), and EinsplineSetBuilder::createSPOSetFromXML().
|
inline |
Definition at line 43 of file createDistanceTable.h.
References createDistanceTableAA(), createDistanceTableAAOMPTarget(), DC_POS_OFFLOAD, and qmcplusplus::Units::time::s.
Referenced by ParticleSet::addTable().
|
inline |
Definition at line 60 of file createDistanceTable.h.
References createDistanceTableAB(), createDistanceTableABOMPTarget(), DC_POS_OFFLOAD, and qmcplusplus::Units::time::s.
| std::unique_ptr< DistanceTable > createDistanceTableAA | ( | ParticleSet & | s, |
| std::ostream & | description | ||
| ) |
Class to manage multiple DistanceTable objects.
Adding SymmetricDTD to the list, e.g., el-el distance table.
free function to create a distable table of s-s
| s | source/target particle set |
Definition at line 27 of file createDistanceTableAA.cpp.
References qmcplusplus::ewaldref::DIM, OHMMS_DIM, OHMMS_PRECISION, qmcplusplus::Units::time::s, SUPERCELL_BULK, SUPERCELL_SLAB, and SUPERCELL_WIRE.
Referenced by createDistanceTable().
| std::unique_ptr< DistanceTable > createDistanceTableAAOMPTarget | ( | ParticleSet & | s, |
| std::ostream & | description | ||
| ) |
Adding SymmetricDTD to the list, e.g., el-el distance table.
| s | source/target particle set |
Definition at line 27 of file createDistanceTableAAOMPTarget.cpp.
References qmcplusplus::ewaldref::DIM, OHMMS_DIM, OHMMS_PRECISION, qmcplusplus::Units::time::s, SUPERCELL_BULK, SUPERCELL_SLAB, and SUPERCELL_WIRE.
Referenced by createDistanceTable().
| std::unique_ptr< DistanceTable > createDistanceTableAB | ( | const ParticleSet & | s, |
| ParticleSet & | t, | ||
| std::ostream & | description | ||
| ) |
free function create a distable table of s-t
Adding AsymmetricDTD to the list, e.g., el-el distance table.
| s | source/target particle set |
Definition at line 27 of file createDistanceTableAB.cpp.
References qmcplusplus::ewaldref::DIM, ParticleSet::getLattice(), OhmmsElementBase::getName(), OHMMS_DIM, qmcplusplus::Units::time::s, SUPERCELL_BULK, SUPERCELL_SLAB, and SUPERCELL_WIRE.
Referenced by createDistanceTable().
| std::unique_ptr< DistanceTable > createDistanceTableABOMPTarget | ( | const ParticleSet & | s, |
| ParticleSet & | t, | ||
| std::ostream & | description | ||
| ) |
Adding AsymmetricDTD to the list, e.g., el-el distance table.
| s | source/target particle set |
Definition at line 27 of file createDistanceTableABOMPTarget.cpp.
References qmcplusplus::ewaldref::DIM, ParticleSet::getLattice(), OhmmsElementBase::getName(), OHMMS_DIM, qmcplusplus::Units::time::s, SUPERCELL_BULK, SUPERCELL_SLAB, and SUPERCELL_WIRE.
Referenced by createDistanceTable().
| DriftModifierBase * createDriftModifier | ( | xmlNodePtr | cur, |
| const Communicate * | myComm | ||
| ) |
create DriftModifier
Definition at line 19 of file DriftModifierBuilder.cpp.
References ParameterSet::add(), Communicate::barrier_and_abort(), and ParameterSet::put().
Referenced by QMCDriver::process(), and QMCDriverNew::QMCDriverNew().
| DriftModifierBase * createDriftModifier | ( | const std::string & | drift_modifier_str, |
| QMCTraits::RealType | unr_a | ||
| ) |
Definition at line 31 of file DriftModifierBuilder.cpp.
References lowerCase().
| std::unique_ptr< DynamicCoordinates > createDynamicCoordinates | ( | const DynamicCoordinateKind | kind | ) |
create DynamicCoordinates based on kind
Definition at line 21 of file DynamicCoordinatesBuilder.cpp.
References DC_POS, and DC_POS_OFFLOAD.
| std::unique_ptr< ParticleSet > createElectronParticleSet | ( | const SimulationCell & | simulation_cell | ) |
Definition at line 24 of file test_hamiltonian_factory.cpp.
References SpeciesSet::addAttribute(), and SpeciesSet::addSpecies().
Referenced by TEST_CASE().
| NewTimer & createGlobalTimer | ( | const std::string & | myname, |
| timer_levels | mylevel | ||
| ) |
Definition at line 51 of file TimerManager.cpp.
References getGlobalTimerManager().
Referenced by TrialWaveFunction::addComponent(), QMCHamiltonian::addOperator(), applyCuspCorrection(), EinsplineSpinorSetBuilder::createSPOSetFromXML(), EinsplineSetBuilder::createSPOSetFromXML(), qmcplusplus::ewaldref::ewaldEnergy(), QMCMain::execute(), generateCuspInfo(), and QMCMain::runQMC().
| std::unique_ptr<OneDimCubicSpline<T> > qmcplusplus::createSpline4RbyVs_temp | ( | const LRHandlerBase * | aLR, |
| T | rcut, | ||
| const LinearGrid< T > & | agrid | ||
| ) |
Definition at line 137 of file LRCoulombSingleton.cpp.
References LRHandlerBase::evaluate(), LinearGrid< T, CT >::makeClone(), and OneDimGridBase< T, CT >::size().
Referenced by LRCoulombSingleton::createSpline4RbyVs().
| std::unique_ptr<OneDimCubicSpline<T> > qmcplusplus::createSpline4RbyVsDeriv_temp | ( | const LRHandlerBase * | aLR, |
| T | rcut, | ||
| const LinearGrid< T > & | agrid | ||
| ) |
Definition at line 161 of file LRCoulombSingleton.cpp.
References LRHandlerBase::evaluate(), LinearGrid< T, CT >::makeClone(), OneDimGridBase< T, CT >::size(), and LRHandlerBase::srDf().
Referenced by LRCoulombSingleton::createSpline4RbyVsDeriv().
| sycl::queue createSYCLInOrderQueueOnDefaultDevice | ( | ) |
create an in-order queue using the default device
Definition at line 20 of file SYCLruntime.cpp.
References getSYCLDefaultDeviceDefaultQueue(), and queue.
Referenced by DelayedUpdateSYCL< T, T_FP >::DelayedUpdateSYCL().
| sycl::queue createSYCLQueueOnDefaultDevice | ( | ) |
create a out-of-order queue using the default device
Definition at line 26 of file SYCLruntime.cpp.
References getSYCLDefaultDeviceDefaultQueue(), and queue.
| WalkerControlBase * createWalkerController | ( | int | nwtot, |
| Communicate * | comm, | ||
| xmlNodePtr | cur, | ||
| bool | reconfig = false |
||
| ) |
function to create WalkerControlBase or its derived class
| current | number of walkers |
set of parameters
Definition at line 28 of file WalkerControlFactory.cpp.
References ParameterSet::add(), app_log(), comm, omp_get_max_threads(), ParameterSet::put(), WalkerControlBase::set_method(), WalkerControlBase::setMinMax(), and Communicate::size().
Referenced by SimpleFixedNodeBranch::initWalkerController(), and SimpleFixedNodeBranch::resetRun().
|
inline |
Definition at line 200 of file TinyVector.h.
Referenced by OTCross< TinyVector< T1, 3 >, TinyVector< T2, 3 > >::apply(), and LatticeAnalyzer< T, 3 >::calcSimulationCellRadius().
| std::atomic<size_t> qmcplusplus::CUDAallocator_device_mem_allocated | ( | 0 | ) |
| qmcplusplus::cudaErrorCheck | ( | cudaMemcpyAsync(dev_lu.data(), lu.data(), sizeof(decltype(lu)::value_type) *lu.size(), cudaMemcpyHostToDevice, hstream) | , |
| "cudaMemcpyAsync failed copying log_values to device" | |||
| ) |
Referenced by qmcplusplus::compute::add_delay_list_save_sigma_VGL_batched(), CUDAManagedAllocator< T >::allocate(), OMPallocator< Value >::allocate(), CUDAAllocator< T_FP >::allocate(), CUDAHostAllocator< T_FP >::allocate(), CUDALockedPageAllocator< T, ULPHA >::allocate(), qmcplusplus::compute::applyW_batched(), qmcplusplus::compute::calcGradients_batched(), qmcplusplus::compute::BLAS::copy_batched(), qmcplusplus::compute::copyAinvRow_saveGL_batched(), CUDAAllocator< T_FP >::copyDeviceToDevice(), CUDAAllocator< T_FP >::copyFromDevice(), CUDAAllocator< T_FP >::copyToDevice(), CUDADeviceManager::CUDADeviceManager(), CUDAfill_n(), CUDAHandles::CUDAHandles(), cuSolverInverter< T_FP >::cuSolverInverter(), CUDAManagedAllocator< T >::deallocate(), OMPallocator< Value >::deallocate(), CUDAAllocator< T_FP >::deallocate(), CUDAHostAllocator< T_FP >::deallocate(), CUDALockedPageAllocator< T, ULPHA >::deallocate(), Queue< PlatformKind::CUDA >::enqueueD2H(), Queue< PlatformKind::CUDA >::enqueueH2D(), qmcplusplus::compute::BLAS::gemv_batched(), qmcplusplus::compute::BLAS::ger_batched(), DelayedUpdateCUDA< T, T_FP >::getInvRow(), DelayedUpdateCUDA< T, T_FP >::initializeInv(), cuSolverInverter< T_FP >::invert_transpose(), rocSolverInverter< T_FP >::invert_transpose(), DiracMatrixComputeCUDA< VALUE_FP >::invert_transpose(), DiracMatrixComputeCUDA< VALUE_FP >::mw_computeInvertAndLog(), DiracMatrixComputeCUDA< VALUE_FP >::mw_computeInvertAndLog_stride(), DiracMatrixComputeCUDA< VALUE_FP >::mw_invertTranspose(), Queue< PlatformKind::CUDA >::Queue(), rocSolverInverter< T_FP >::rocSolverInverter(), ScopedProfiler::ScopedProfiler(), Queue< PlatformKind::CUDA >::sync(), TEST_CASE(), DelayedUpdateCUDA< T, T_FP >::updateInvMat(), CUDAHandles::~CUDAHandles(), cuSolverInverter< T_FP >::~cuSolverInverter(), Queue< PlatformKind::CUDA >::~Queue(), rocSolverInverter< T_FP >::~rocSolverInverter(), and ScopedProfiler::~ScopedProfiler().
| qmcplusplus::cudaErrorCheck | ( | cudaMemcpyAsync(dev_lu2.data(), lu2.data(), sizeof(decltype(lu2)::value_type) *lu2.size(), cudaMemcpyHostToDevice, hstream) | , |
| "cudaMemcpyAsync failed copying log_values to device" | |||
| ) |
| qmcplusplus::cudaErrorCheck | ( | cudaMemcpyAsync(dev_lus.data(), lus.data(), sizeof(decltype(lus)::value_type) *lus.size(), cudaMemcpyHostToDevice, hstream) | , |
| "cudaMemcpyAsync failed copying log_values to device" | |||
| ) |
| qmcplusplus::cudaErrorCheck | ( | cudaMemcpyAsync(dev_pivots.data(), pivots.data(), sizeof(int) *pivots.size(), cudaMemcpyHostToDevice, hstream) | , |
| "cudaMemcpyAsync failed copying log_values to device" | |||
| ) |
| qmcplusplus::cudaErrorCheck | ( | cudaMemcpyAsync(log_values.data(), dev_log_values.data(), sizeof(std::complex< double >) *2, cudaMemcpyDeviceToHost, hstream) | , |
| "cudaMemcpyAsync failed copying log_values from device" | |||
| ) |
| cudaErrorCheck | ( | cudaStreamSynchronize(hstream) | , |
| "cudaStreamSynchronize failed!" | |||
| ) |
| qmcplusplus::cudaErrorCheck | ( | cudaMemcpyAsync(devM_vec.data(), M_vec.data(), sizeof(decltype(M_vec)::value_type) *M_vec.size(), cudaMemcpyHostToDevice, hstream) | , |
| "cudaMemcpyAsync failed copying M to device" | |||
| ) |
| qmcplusplus::cudaErrorCheck | ( | cudaMemcpyAsync(devM2_vec.data(), M2_vec.data(), sizeof(decltype(M2_vec)::value_type) *M2_vec.size(), cudaMemcpyHostToDevice, hstream) | , |
| "cudaMemcpyAsync failed copying M2 to device" | |||
| ) |
| qmcplusplus::cudaErrorCheck | ( | cudaMemcpyAsync(devMs.data(), Ms.data(), sizeof(decltype(Ms)::value_type) *Ms.size(), cudaMemcpyHostToDevice, hstream) | , |
| "cudaMemcpyAsync failed copying Ms to device" | |||
| ) |
| qmcplusplus::cudaErrorCheck | ( | cudaMemcpyAsync(M_vec.data(), devM_vec.data(), sizeof(decltype(M_vec)::value_type) *M_vec.size(), cudaMemcpyDeviceToHost, hstream) | , |
| "cudaMemcpyAsync failed copying invM from device" | |||
| ) |
| qmcplusplus::cudaErrorCheck | ( | cudaMemcpyAsync(M2_vec.data(), devM2_vec.data(), sizeof(decltype(M2_vec)::value_type) *M2_vec.size(), cudaMemcpyDeviceToHost, hstream) | , |
| "cudaMemcpyAsync failed copying invM from device" | |||
| ) |
| qmcplusplus::cudaErrorCheck | ( | cudaMemcpyAsync(pivots.data(), dev_pivots.data(), sizeof(int) *pivots.size(), cudaMemcpyDeviceToHost, hstream) | , |
| "cudaMemcpyAsync failed copying pivots from device" | |||
| ) |
| void CUDAfill_n | ( | T * | ptr, |
| size_t | n, | ||
| const T & | value | ||
| ) |
fill device memory with a given value.
| ptr | pointer to device memory |
| n | size of type T elemements desired value. Due to cudaMemset limitation. Only filling 0 is supported. |
this function is only intended to prevent NaN in containers when device memory segments are allocated. do not use outside allocators.
Definition at line 20 of file CUDAfill.cpp.
References cudaErrorCheck(), cudaMemset, and n.
Referenced by qmc_allocator_traits< qmcplusplus::CUDAAllocator< T > >::fill_n().
| template void CUDAfill_n< double > | ( | double * | ptr, |
| size_t | n, | ||
| const double & | value | ||
| ) |
| template void CUDAfill_n< float > | ( | float * | ptr, |
| size_t | n, | ||
| const float & | value | ||
| ) |
| template void CUDAfill_n< int > | ( | int * | ptr, |
| size_t | n, | ||
| const int & | value | ||
| ) |
| template void CUDAfill_n< size_t > | ( | size_t * | ptr, |
| size_t | n, | ||
| const size_t & | value | ||
| ) |
| template void CUDAfill_n< std::complex< double > > | ( | std::complex< double > * | ptr, |
| size_t | n, | ||
| const std::complex< double > & | value | ||
| ) |
| template void CUDAfill_n< std::complex< float > > | ( | std::complex< float > * | ptr, |
| size_t | n, | ||
| const std::complex< float > & | value | ||
| ) |
|
inline |
delete the pointers in [first,last)
Definition at line 22 of file IteratorUtility.h.
Referenced by SPOSetInfo::clear(), SPOSetInfoSimple< qmcplusplus::SHOState >::clear(), DensityMatrices1B::finalize(), TraceSamples< std::complex< TraceReal > >::finalize(), QMCCostFunction::getConfigurations(), QMCCostFunctionBatched::getConfigurations(), CloneManager::~CloneManager(), CSUpdateBase::~CSUpdateBase(), EnergyDensityEstimator::~EnergyDensityEstimator(), QMCCostFunction::~QMCCostFunction(), QMCCostFunctionBase::~QMCCostFunctionBase(), SPOSetInfo::~SPOSetInfo(), and SPOSetInfoSimple< qmcplusplus::SHOState >::~SPOSetInfoSimple().
|
inline |
calculate the derivative of a Ylm with respect to theta and phi param[in] l: angular momentum param[in] m: magnetic quantum number param[in] r: cartesian position align with [z,x,y].
note: MUST BE UNIT VECTOR to be consistent with Ylm above param[out] theta_deriv: derivative of Ylm with respect to theta param[out] phi_deriv: derivative of Ylm with respect to phi param[in] conj: true if we are taking derivatives of conj(Ylm)
Definition at line 115 of file Ylm.h.
References acos(), atan2(), conj(), cos(), qmcplusplus::Units::distance::m, sin(), sqrt(), tan(), and Ylm().
Referenced by sphericalHarmonicGrad(), and TEST_CASE().
Definition at line 838 of file TensorOps.h.
Referenced by DiracDeterminant< DU_TYPE >::acquireResource(), MultiDiracDeterminant::acquireResource(), DiracDeterminantBatched< PL, VT, FPVT >::acquireResource(), EinsplineSetBuilder::AnalyzeTwists2(), SlaterDetBuilder::buildComponent(), WaveFunctionTester::checkGradientAtConfiguration(), MultiSlaterDetTableMethod::createResource(), MultiSlaterDetTableMethod::evaluateMultiDiracDeterminantDerivatives(), MultiSlaterDetTableMethod::evaluateMultiDiracDeterminantDerivativesWF(), qmcplusplus::ewaldref::ewaldSum(), ExpFitClass< M >::Fit(), ExpFitClass< M >::FitCusp(), EshdfFile::getPtvs(), SpaceGrid::initialize_rectilinear(), inverse(), SlaterDetWithBackflow::isOptimizable(), SlaterDet::isOptimizable(), qmcplusplus::ewaldref::madelungSum(), SlaterDetWithBackflow::makeClone(), MultiSlaterDetTableMethod::makeClone(), SlaterDet::makeClone(), MultiSlaterDetTableMethod::mw_accept_rejectMove(), DiracDeterminantBatched< PL, VT, FPVT >::mw_accept_rejectMove(), DiracDeterminantBatched< PL, VT, FPVT >::mw_calcRatio(), MultiSlaterDetTableMethod::mw_calcRatio(), DiracDeterminantBatched< PL, VT, FPVT >::mw_completeUpdates(), DiracDeterminantBatched< PL, VT, FPVT >::mw_evalGrad(), MultiSlaterDetTableMethod::mw_evalGrad_impl(), DiracDeterminantBatched< PL, VT, FPVT >::mw_evalGradWithSpin(), MultiDiracDeterminant::mw_evaluateDetsAndGradsForPtclMove(), MultiDiracDeterminant::mw_evaluateDetsForPtclMove(), DiracDeterminantBatched< PL, VT, FPVT >::mw_evaluateGL(), MultiDiracDeterminant::mw_evaluateGrads(), DiracDeterminantBatched< PL, VT, FPVT >::mw_evaluateLog(), DiracDeterminant< DU_TYPE >::mw_evaluateRatios(), DiracDeterminantBatched< PL, VT, FPVT >::mw_evaluateRatios(), DiracDeterminantBatched< PL, VT, FPVT >::mw_invertPsiM(), MultiSlaterDetTableMethod::mw_prepareGroup(), DiracDeterminant< DU_TYPE >::mw_ratioGrad(), MultiSlaterDetTableMethod::mw_ratioGrad(), DiracDeterminantBatched< PL, VT, FPVT >::mw_ratioGrad(), DiracDeterminantBatched< PL, VT, FPVT >::mw_ratioGradWithSpin(), DiracDeterminantBatched< PL, VT, FPVT >::mw_recompute(), EinsplineSetBuilder::ReadOrbitalInfo_ESHDF(), DiracDeterminant< DU_TYPE >::releaseResource(), DiracDeterminantBatched< PL, VT, FPVT >::releaseResource(), CrystalLattice< ST, 3 >::reset(), NESpaceGrid< REAL >::someMoreAxisGridStuff(), and TWF().
Definition at line 848 of file TensorOps.h.
Definition at line 857 of file TensorOps.h.
Definition at line 866 of file TensorOps.h.
|
inline |
Definition at line 89 of file determinant_operators.h.
References BLAS::cone, BLAS::copy(), BLAS::gemv(), BLAS::ger(), and qmcplusplus::Units::distance::m.
Referenced by InverseUpdateByColumn().
|
inline |
Definition at line 72 of file determinant_operators.h.
References BLAS::gemv(), BLAS::ger(), and qmcplusplus::Units::distance::m.
Referenced by InverseUpdateByRow().
|
inline |
determinant of a matrix
| x | starting address of an n-by-m matrix |
| n | rows |
| m | cols |
| pivot | integer pivot array |
Definition at line 141 of file DeterminantOperators.h.
References LUFactorization(), qmcplusplus::Units::distance::m, and n.
Referenced by SmallMatrixDetCalculator< ValueType >::evaluate().
|
inline |
distribute MPI ranks among devices
the amount of MPI ranks for each device differs by 1 at maximum. larger id has more MPI ranks.
Definition at line 24 of file determineDefaultDeviceNum.h.
References num_ranks.
Referenced by CUDADeviceManager::CUDADeviceManager(), OMPDeviceManager::OMPDeviceManager(), and SYCLDeviceManager::SYCLDeviceManager().
|
inline |
determinant ratio with a column substitution
| Minv | inverse matrix |
| newv | column vector |
| colchanged | column index to be replaced |
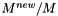
Definition at line 232 of file DeterminantOperators.h.
References qmcplusplus::simd::dot().
Referenced by MultiDiracDeterminant::evaluateDetsAndGradsForPtclMove(), MultiDiracDeterminant::evaluateDetsAndGradsForPtclMoveWithSpin(), MultiDiracDeterminant::evaluateDetsForPtclMove(), AGPDeterminant::ratio(), and AGPDeterminant::ratioDown().
|
inline |
determinant ratio with a row substitution
| Minv | inverse matrix |
| newv | row vector |
| rowchanged | row index to be replaced |
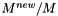
Definition at line 219 of file DeterminantOperators.h.
References qmcplusplus::simd::dot().
Referenced by AGPDeterminant::ratio(), and AGPDeterminant::ratioUp().
| std::vector<int, CUDAAllocator<int> > qmcplusplus::dev_infos | ( | pivots. | size() | ) |
Referenced by TEST_CASE().
| std::vector<StdComp, CUDAAllocator<StdComp> > qmcplusplus::dev_log_values | ( | batch_size | ) |
Referenced by TEST_CASE().
| std::vector<StdComp, CUDAHostAllocator<StdComp> > qmcplusplus::dev_lu | ( | lu. | size() | ) |
Referenced by TEST_CASE().
| std::vector<StdComp, CUDAHostAllocator<StdComp> > qmcplusplus::dev_lu2 | ( | lu2. | size() | ) |
| std::vector<StdComp*, CUDAAllocator<StdComp*> > qmcplusplus::dev_lus | ( | batch_size | ) |
Referenced by TEST_CASE().
| std::vector< int, CUDAAllocator< int > > dev_pivots | ( | pivots. | size() | ) |
Referenced by TEST_CASE().
| std::vector<double, CUDAAllocator<double> > qmcplusplus::devM2_vec | ( | M2_vec. | size() | ) |
| std::vector<double, CUDAAllocator<double> > qmcplusplus::devM_vec | ( | M_vec. | size() | ) |
Referenced by TEST_CASE().
| std::vector<double*, CUDAAllocator<double*> > qmcplusplus::devMs | ( | Ms. | size() | ) |
Referenced by TEST_CASE().
|
inline |
Convenience function to compute 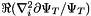 .
.
| g | OHMMS_DIM dimensional vector for 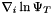 . . |
| l | A number, representing 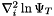 . . |
| gg | OHMMS_DIM dimensional vector containing 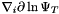 . . |
| gl | A number, representing 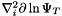 |
| ideriv | A number, representing 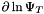 |
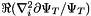
Definition at line 51 of file BareKineticHelper.h.
References dot().
Referenced by BareKineticEnergy::evaluateWithIonDerivs().
|
inline |
Definition at line 57 of file BareKineticHelper.h.
References OTCDot< T1, T2, D >::apply().
| template void qmcplusplus::DMCBatched::advanceWalkers< CoordsType::POS > | ( | const StateForThread & | sft, |
| Crowd & | crowd, | ||
| DriverTimers & | timers, | ||
| DMCTimers & | dmc_timers, | ||
| ContextForSteps & | step_context, | ||
| bool | recompute, | ||
| bool | accumulate_this_step | ||
| ) |
| template void qmcplusplus::DMCBatched::advanceWalkers< CoordsType::POS_SPIN > | ( | const StateForThread & | sft, |
| Crowd & | crowd, | ||
| DriverTimers & | timers, | ||
| DMCTimers & | dmc_timers, | ||
| ContextForSteps & | step_context, | ||
| bool | recompute, | ||
| bool | accumulate_this_step | ||
| ) |
| void qmcplusplus::doSOECPotentialTest | ( | bool | use_VPs | ) |
Definition at line 75 of file test_SOECPotential.cpp.
References SpeciesSet::addAttribute(), SOECPotential::addComponent(), TrialWaveFunction::addComponent(), SpeciesSet::addSpecies(), TestSOECPotential::addVPs(), Matrix< T, Alloc >::begin(), RadialJastrowBuilder::buildComponent(), CHECK(), Matrix< T, Alloc >::cols(), OHMMS::Controller, TestSOECPotential::copyGridUnrotatedForTest(), ParticleSet::create(), SOECPotential::createResource(), TrialWaveFunction::createResource(), ParticleSet::createSK(), TestSOECPotential::didGridChange(), doc, TestSOECPotential::evalFast(), qmcplusplus::testing::getParticularListener(), Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), ParticleSet::getTotalNum(), lattice, SOECPotential::makeClone(), TrialWaveFunction::makeClone(), TestSOECPotential::mw_evaluateImpl(), TrialWaveFunction::mw_evaluateLog(), ParticleSet::mw_update(), qmcplusplus::hdf::num_walkers, okay, OMPTARGET, Libxml2Document::parseFromString(), ECPComponentBuilder::pp_so, ParticleSet::R, ECPComponentBuilder::read_pp_file(), REQUIRE(), ParticleSet::resetGroups(), ParticleSet::setName(), SOECPotential::setRandomGenerator(), ParticleSet::setSpinor(), ParticleSet::spins, and ParticleSet::update().
Referenced by TEST_CASE().
|
inline |
Definition at line 122 of file ParticleAttribOps.h.
References dot().
Referenced by DMCUpdateAllWithRejection::advanceWalker(), VMCUpdateAll::advanceWalker(), DMCUpdateAllWithKill::advanceWalker(), CSVMCUpdateAllWithDrift::advanceWalker(), RMCUpdateAllWithDrift::advanceWalkersRMC(), RMCUpdateAllWithDrift::advanceWalkersVMC(), complex_test_case(), ACForce::compute_regularizer_f(), double_test_case(), BareKineticEnergy::evaluate(), BareKineticEnergy::evaluate_orig(), DiracDeterminantWithBackflow::evaluateDerivatives(), J1Spin< FT >::evaluateDerivatives(), TwoBodyJastrow< FT >::evaluateDerivatives(), J1OrbitalSoA< FT >::evaluateDerivatives(), BareKineticEnergy::evaluateWithIonDerivs(), getDriftScale(), normalize(), WaveFunctionTester::runZeroVarianceTest(), TEST_CASE(), test_J3_polynomial3D(), and BackflowTransformation::testPbyP().
|
inline |
Definition at line 133 of file ParticleAttribOps.h.
References OTCDot< T1, T2, D >::apply().
|
inline |
Definition at line 190 of file TinyVector.h.
|
inline |
Definition at line 388 of file SymTensor.h.
|
inline |
Definition at line 395 of file SymTensor.h.
|
inline |
Definition at line 402 of file SymTensor.h.
|
inline |
Definition at line 409 of file SymTensor.h.
|
inline |
|
inline |
Definition at line 416 of file SymTensor.h.
|
inline |
|
inline |
Tensor-Vector dot product 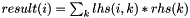 .
.
| lhs | a tensor |
| rhs | a vector |
Definition at line 436 of file Tensor.h.
|
inline |
Definition at line 520 of file AntiSymTensor.h.
Referenced by MomentumDistribution::accumulate(), NESpaceGrid< REAL >::accumulate(), SODMCUpdatePbyPWithRejectionFast::advanceWalker(), DMCUpdatePbyPWithRejectionFast::advanceWalker(), SOVMCUpdatePbyP::advanceWalker(), VMCUpdatePbyP::advanceWalker(), DMCUpdatePbyPL2::advanceWalker(), DMCBatched::advanceWalkers(), RMCUpdatePbyPWithDrift::advanceWalkersRMC(), RMCUpdatePbyPWithDrift::advanceWalkersVMC(), EinsplineSetBuilder::AnalyzeTwists2(), DTD_BConds< T, 3, PPPO >::apply_bc(), DTD_BConds< T, D, SC >::apply_bc(), DTD_BConds< T, 3, PPPG >::apply_bc(), DTD_BConds< T, 2, PPPO >::apply_bc(), DTD_BConds< T, 2, SUPERCELL_WIRE >::apply_bc(), DTD_BConds< T, 3, PPNG >::apply_bc(), DTD_BConds< T, 3, PPNO >::apply_bc(), DTD_BConds< T, 3, SUPERCELL_WIRE >::apply_bc(), CrystalLattice< ST, 3 >::applyMinimumImage(), KContainer::BuildKLists(), LatticeAnalyzer< T, 3 >::calcSimulationCellRadius(), LatticeAnalyzer< T, 2 >::calcSimulationCellRadius(), LatticeAnalyzer< T, 3 >::calcSolidAngles(), LatticeAnalyzer< T, 2 >::calcSolidAngles(), SpinDensityInput::calculateDerivedParameters(), MagnetizationDensityInput::calculateDerivedParameters(), NonLocalECPComponent::calculateProjector(), LatticeAnalyzer< T, 3 >::calcWignerSeitzRadius(), LatticeAnalyzer< T, 2 >::calcWignerSeitzRadius(), SpaceGrid::check_grid(), NESpaceGrid< REAL >::check_grid(), BsplineReader::check_twists(), DiracDeterminantBase::checkG(), QMCUpdateBase::checkLogAndGL(), QMCDriverNew::checkLogAndGL(), NaNguard::checkOneParticleGradients(), DiracDeterminantBatched< PL, VT, FPVT >::computeGL(), QMCDriverNew::computeLogGreensFunction(), StructFact::computeRhok(), kSpaceJastrow::copyFromBuffer(), SplineR2R< ST >::create_spline(), CountingGaussian::d_to_b(), dlaplacian(), Dot(), CrystalLattice< ST, 3 >::Dot(), double_test_case(), DiracDeterminantWithBackflow::dummyEvalLi(), kSpaceJastrow::Equivalent(), DiracDeterminantWithBackflow::evalGrad(), kSpaceJastrow::evalGrad(), LCAOrbitalBuilder::EvalPeriodicImagePhaseFactors(), MomentumEstimator::evaluate(), SpaceGrid::evaluate(), HarmonicExternalPotential::evaluate(), CountingGaussianRegion::evaluate(), PWBasis::evaluate(), CountingGaussian::evaluate(), Gvectors< ST, LT >::evaluate_KE(), FreeOrbital::evaluate_notranspose(), CountingGaussian::evaluate_print(), Gvectors< ST, LT >::evaluate_psi_r(), DTD_BConds< T, D, SC >::evaluate_rsquared(), GridExternalPotential::evaluate_sp(), HarmonicExternalPotential::evaluate_sp(), BareKineticEnergy::evaluate_sp(), CountingGaussian::evaluateDerivative_A(), CountingGaussian::evaluateDerivative_B(), DiracDeterminantWithBackflow::evaluateDerivatives(), RotatedSPOs::evaluateDerivatives(), MultiSlaterDetTableMethod::evaluateDerivatives(), kSpaceJastrow::evaluateDerivatives(), CountingGaussianRegion::evaluateDerivatives(), CountingJastrow< RegionType >::evaluateDerivatives(), JeeIOrbitalSoA< FT >::evaluateDerivatives(), CountingJastrow< RegionType >::evaluateExponents(), DiracDeterminantWithBackflow::evaluateLog(), MultiSlaterDetTableMethod::evaluateLog(), kSpaceJastrow::evaluateLog(), DiracDeterminant< DU_TYPE >::evaluateLog(), CountingGaussian::evaluateLog(), AGPDeterminant::evaluateLogAndStore(), BareKineticEnergy::evaluateOneBodyOpMatrix(), NonLocalECPComponent::evaluateOneBodyOpMatrixContribution(), NonLocalECPComponent::evaluateOneBodyOpMatrixdRContribution(), BareKineticEnergy::evaluateOneBodyOpMatrixForceDeriv(), NonLocalECPComponent::evaluateOneWithForces(), BackflowTransformation::evaluatePbyP(), BackflowTransformation::evaluatePbyPAll(), BackflowTransformation::evaluatePbyPWithGrad(), kSpaceJastrow::evaluateRatiosAlltoOne(), CountingGaussianRegion::evaluateTemp(), CountingJastrow< RegionType >::evaluateTempExponents(), SoaAtomicBasisSet< ROT, SH >::evaluateV(), FreeOrbital::evaluateValue(), NonLocalECPComponent::evaluateValueAndDerivatives(), SoaAtomicBasisSet< ROT, SH >::evaluateVGH(), SoaAtomicBasisSet< ROT, SH >::evaluateVGHGH(), FreeOrbital::evaluateVGL(), SoaAtomicBasisSet< ROT, SH >::evaluateVGL(), qmcplusplus::ewaldref::ewaldEnergy(), extrap(), SkPot::FillFk(), KContainer::findApproxMMax(), found_shorter_base(), FreeOrbital::FreeOrbital(), DensityMatrices1B::generate_density_samples(), OneBodyDensityMatrices::generateDensitySamples(), OneBodyDensityMatrices::generateDensitySamplesWithSpin(), DriftModifierUNR::getDrift(), getScaledDrift(), getScaledDriftL2(), Gvectors< ST, LT >::Gvectors(), MPC::init_f_G(), MPC::init_gvecs(), MPC::init_spline(), SpaceGrid::initialize_rectilinear(), HybridRepCenterOrbitals< SPLINEBASE::DataType >::interpolate_buffer_vgl(), CrystalLattice< ST, 3 >::k_cart(), CountingGaussian::k_to_c(), CrystalLattice< ST, 3 >::k_unit(), kSpaceJastrow::kSpaceJastrow(), CrystalLattice< ST, 3 >::ksq(), laplacian(), QMCUpdateBase::logBackwardGF(), MomentumDistribution::MomentumDistribution(), BareKineticEnergy::mw_evaluatePerParticle(), NEReferencePoints::NEReferencePoints(), RspaceMadelungTerm::operator()(), KspaceMadelungTerm::operator()(), RspaceEwaldTerm::operator()(), KspaceEwaldTerm::operator()(), magLess::operator()(), kSpaceJastrow::operator()(), ReferencePoints::put(), SpinDensity::put(), MomentumEstimator::putSpecial(), Quadrature3D< T >::Quadrature3D(), kSpaceJastrow::ratio(), DiracDeterminantWithBackflow::ratioGrad(), kSpaceJastrow::ratioGrad(), EinsplineSetBuilder::ReadOrbitalInfo_ESHDF(), CrystalLattice< ST, 3 >::reset(), SplineC2R< ST >::resize_kpoints(), SplineC2C< ST >::resize_kpoints(), SplineC2COMPTarget< ST >::resize_kpoints(), SplineC2ROMPTarget< ST >::resize_kpoints(), SOECPComponent::rotateQuadratureGrid(), NonLocalECPComponent::rotateQuadratureGrid(), EinsplineSetBuilder::set_metadata(), LRBreakupParameters< T, 3 >::SetLRCutoffs(), setScaledDriftPbyPandNodeCorr(), PWBasis::setTwistAngle(), kSpaceJastrow::setupGvecs(), NESpaceGrid< REAL >::someMoreAxisGridStuff(), kSpaceJastrow::StructureFactor(), RotatedSPOs::table_method_eval(), TEST_CASE(), test_tiny_vector(), CrystalLattice< ST, 3 >::toCart(), CrystalLattice< ST, 3 >::toUnit(), PWBasis::trimforecut(), DiracDeterminant< DU_TYPE >::updateAfterSweep(), and MultiSlaterDetTableMethod::updateBuffer().
|
inline |
Definition at line 527 of file AntiSymTensor.h.
|
inline |
Definition at line 534 of file AntiSymTensor.h.
|
inline |
Definition at line 541 of file AntiSymTensor.h.
|
inline |
Definition at line 548 of file AntiSymTensor.h.
|
inline |
Definition at line 555 of file AntiSymTensor.h.
|
inline |
Definition at line 562 of file AntiSymTensor.h.
|
inline |
Definition at line 158 of file ParticleAttribOps.h.
References OTCDot_CC< T1, T2, D >::apply().
Referenced by complex_test_case(), and ACForce::compute_regularizer_f().
| void qmcplusplus::double_test_case | ( | ) |
Definition at line 31 of file test_attrib_ops.cpp.
References CHECK(), Dot(), dot(), REQUIRE(), Vector< T, Alloc >::resize(), and Vector< T, Alloc >::size().
| std::atomic<size_t> qmcplusplus::dual_device_mem_allocated | ( | 0 | ) |
| testing::EstimatorManagerNewTest qmcplusplus::embt | ( | ham | , |
| c | , | ||
| num_ranks | |||
| ) |
| EstimatorManagerInput qmcplusplus::emi | ( | estimators_doc. | getRoot() | ) |
| EstimatorManagerInput qmcplusplus::emi2 | ( | estimators_doc2. | getRoot() | ) |
estimate equilibration of block data
Assumes vals is an array to be averaged. This will estimate which, if any, of the data should be thrown out as equilibration data. Returns the index of the array to start averaging from. frac is the fraction of the data to average initially to help determine the equilibration
Definition at line 50 of file FSUtilities.cpp.
References getStats().
Referenced by SkParserHDF5::parse().
|
inline |
Definition at line 398 of file OhmmsVector.h.
References forEach(), and Vector< T, Alloc >::size().
|
inline |
Definition at line 514 of file OhmmsMatrix.h.
References Matrix< T, Alloc >::cols(), forEach(), and Matrix< T, Alloc >::rows().
Referenced by assign(), calcSmallDeterminant(), CustomizedMatrixDet< 5 >::evaluate(), Hdispatcher::flex_evaluate(), ParticleSet::loadWalker(), ParticleSet::makeMoveAllParticles(), ParticleSet::makeMoveAllParticlesWithDrift(), MultiDiracDeterminant::mw_updateRatios(), operator &=(), operator%=(), operator*=(), operator+=(), operator-=(), operator/=(), operator<<=(), operator>>=(), operator^=(), operator|=(), and ParticleSet::update().
|
inline |
Definition at line 896 of file ParticleAttrib.cpp.
References forEach(), and Vector< T, Alloc >::size().
| RealType qmcplusplus::evaluateForPhi0Body | ( | RealType | phi0, |
| ValueVector & | pos, | ||
| ValueVector & | ELcurr, | ||
| ValueVector & | ELideal, | ||
| CuspCorrection & | cusp, | ||
| OneMolecularOrbital & | phiMO, | ||
| ValGradLap | phiAtRc, | ||
| RealType | etaAtZero, | ||
| RealType | ELorigAtRc, | ||
| RealType | Z | ||
| ) |
Definition at line 453 of file CuspCorrectionConstruction.cpp.
References CuspCorrectionParameters::alpha, CuspCorrectionParameters::C, CuspCorrection::cparam, evalX(), getCurrentLocalEnergy(), getELchi2(), getZeff(), ValGradLap::grad, ValGradLap::lap, phiBar(), CuspCorrectionParameters::Rc, CuspCorrectionParameters::sg, ValGradLap::val, and X2alpha().
Referenced by minimizeForPhiAtZero().
| void evalX | ( | RealType | valRc, |
| GradType | gradRc, | ||
| ValueType | lapRc, | ||
| RealType | Rc, | ||
| RealType | Z, | ||
| RealType | C, | ||
| RealType | valAtZero, | ||
| RealType | eta0, | ||
| TinyVector< ValueType, 5 > & | X | ||
| ) |
Evaluate various orbital quantities that enter as constraints on the correction.
| valRc | orbital value at Rc |
| gradRc | orbital gradient at Rc |
| lapRc | orbital laplacian at Rc |
| Rc | cutoff radius |
| Z | nuclear charge |
| C | offset to keep correction to a single sign |
| valAtZero | orbital value at zero |
| eta0 | value of non-corrected pieces of the orbital at zero |
| X | output |
Definition at line 330 of file CuspCorrectionConstruction.cpp.
References abs(), qmcplusplus::Units::charge::C, and log().
Referenced by evaluateForPhi0Body().
|
inline |
Definition at line 80 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by SelfHealingOverlap::accumulate(), SOVMCUpdateAll::advanceWalker(), DMCUpdateAllWithRejection::advanceWalker(), SODMCUpdatePbyPWithRejectionFast::advanceWalker(), VMCUpdateAll::advanceWalker(), DMCUpdatePbyPWithRejectionFast::advanceWalker(), SOVMCUpdatePbyP::advanceWalker(), VMCUpdatePbyP::advanceWalker(), DMCUpdatePbyPL2::advanceWalker(), CSVMCUpdateAll::advanceWalker(), DMCUpdateAllWithKill::advanceWalker(), CSVMCUpdateAllWithDrift::advanceWalker(), VMCBatched::advanceWalkers(), DMCBatched::advanceWalkers(), RMCUpdatePbyPWithDrift::advanceWalkersRMC(), RMCUpdateAllWithDrift::advanceWalkersRMC(), RMCUpdatePbyPWithDrift::advanceWalkersVMC(), RMCUpdateAllWithDrift::advanceWalkersVMC(), AtomicOrbitals< ST >::AtomicOrbitals(), SFNBranch::branchWeight(), SimpleFixedNodeBranch::branchWeight(), SFNBranch::branchWeightBare(), SimpleFixedNodeBranch::branchWeightBare(), SimpleFixedNodeBranch::branchWeightTau(), QMCUpdateBase::checkLogAndGL(), QMCDriverNew::checkLogAndGL(), chid(), chiu(), CSUpdateBase::computeSumRatio(), LogToValue< T >::convert(), LogToValue< std::complex< T > >::convert(), QMCCostFunction::correlatedSampling(), QMCCostFunctionBatched::correlatedSampling(), GaussianCombo< T >::BasicGaussian::df(), YukawaBreakup< T >::df(), GaussianTimesRN< T >::BasicGaussian::df(), GenericSTO< T >::df(), RadialSTO< T >::df(), DerivYukawaBreakup< T >::df(), ExpFitClass< M >::eval(), ComplexExpFitClass< M >::eval(), EwaldHandler3D::evaldYkgstrain(), SelfHealingOverlapLegacy::evaluate(), GaussianCombo< T >::BasicGaussian::evaluate(), GaussianTimesRN< T >::BasicGaussian::evaluate(), GenericSTO< T >::evaluate(), ShortRangeCuspFunctor< T >::evaluate(), RadialSTO< T >::evaluate(), CountingGaussianRegion::evaluate(), CountingGaussian::evaluate(), ScaledPadeFunctor< T >::evaluate(), SHOSet::evaluate_check(), SHOSet::evaluate_d0(), CountingGaussian::evaluate_print(), EwaldHandler3D::evaluate_vlr_k(), ShortRangeCuspFunctor< T >::evaluateDerivatives(), TwoBodyJastrow< FT >::evaluateRatios(), J1Spin< FT >::evaluateRatios(), J1OrbitalSoA< FT >::evaluateRatios(), JeeIOrbitalSoA< FT >::evaluateRatios(), TwoBodyJastrow< FT >::evaluateRatiosAlltoOne(), kSpaceJastrow::evaluateRatiosAlltoOne(), J1Spin< FT >::evaluateRatiosAlltoOne(), JeeIOrbitalSoA< FT >::evaluateRatiosAlltoOne(), J1OrbitalSoA< FT >::evaluateRatiosAlltoOne(), CountingGaussianRegion::evaluateTemp(), EwaldHandler3D::evalYkgstrain(), RotatedSPOs::exponentiate_antisym_matrix(), GaussianTimesRN< T >::BasicGaussian::f(), GaussianCombo< T >::BasicGaussian::f(), GenericSTO< T >::f(), RadialSTO< T >::f(), SkPot::FillFk(), EwaldHandler3D::fillFk(), DensityMatrices1B::generate_density_samples(), OneBodyDensityMatrices::generateDensitySamples(), OneBodyDensityMatrices::generateDensitySamplesWithSpin(), LogGridLight< T >::getCL(), OptimalGrid::Init(), CSUpdateBase::initCSWalkers(), CSUpdateBase::initCSWalkersForPbyP(), OptimalGrid::InitRatio(), EwaldHandler3D::lrDf(), BackflowBuilder::makeShortRange_twoBody(), TwoBodyJastrow< FT >::mw_calcRatio(), TwoBodyJastrow< FT >::mw_evaluateRatios(), J1OrbitalSoA< FT >::mw_evaluateRatios(), TwoBodyJastrow< FT >::mw_ratioGrad(), OneBodyDensityMatrices::OneBodyDensityMatrices(), KspaceMadelungTerm::operator()(), YukawaBreakup< T >::operator()(), FnExp::operator()(), KspaceEwaldTerm::operator()(), DerivYukawaBreakup< T >::operator()(), OptimalGrid2::OptimalGrid2(), orbd(), orbu(), QMCFixedSampleLinearOptimizeBatched::previous_linear_methods_run(), ExampleHeComponent::ratio(), LatticeGaussianProduct::ratio(), kSpaceJastrow::ratio(), TwoBodyJastrow< FT >::ratio(), J1Spin< FT >::ratio(), J1OrbitalSoA< FT >::ratio(), JeeIOrbitalSoA< FT >::ratio(), CountingJastrow< RegionType >::ratio(), ExampleHeComponent::ratioGrad(), LatticeGaussianProduct::ratioGrad(), kSpaceJastrow::ratioGrad(), TwoBodyJastrow< FT >::ratioGrad(), CountingJastrow< RegionType >::ratioGrad(), JeeIOrbitalSoA< FT >::ratioGrad(), J1OrbitalSoA< FT >::ratioGrad(), J1Spin< FT >::ratioGrad(), CuspCorrection::Rr(), QMCFixedSampleLinearOptimize::run(), WaveFunctionTester::runBasicTest(), WaveFunctionTester::runRatioTest(), WaveFunctionTester::runZeroVarianceTest(), LogGridLight< T >::set(), LogGrid< T, CT >::set(), LogGridZero< T, CT >::set(), DensityMatrices1B::set_state(), EwaldHandlerQuasi2D::slab_vsr_k0(), EwaldHandlerQuasi2D::slabFunc(), EwaldHandler3D::srDf(), TEST_CASE(), test_DiracDeterminant_delayed_update(), test_DiracDeterminant_second(), test_DiracDeterminantBatched_delayed_update(), test_DiracDeterminantBatched_second(), TWF(), SHOSetBuilder::update_basis_states(), CSUpdateBase::updateCSWalkers(), DescentEngine::updateParameters(), Wso(), YukawaBreakup< T >::Xk(), DerivYukawaBreakup< T >::Xk(), and Ylm().
|
inline |
Definition at line 1427 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
| auto qmcplusplus::expectedReferencePoints | ( | ) |
| std::string extractCoefficientsID | ( | xmlNodePtr | cur | ) |
return the id of the first coefficients. If not found, return an emtpy string
Definition at line 33 of file OptimizableFunctorBase.cpp.
References getXMLAttributeValue().
Referenced by BackflowBuilder::addOneBody(), BackflowBuilder::addTwoBody(), RadialJastrowBuilder::createJ1(), RadialJastrowBuilder::createJ2(), BackflowBuilder::makeShortRange_twoBody(), and eeI_JastrowBuilder::putkids().
|
inline |
Definition at line 129 of file MPC.cpp.
References qmcplusplus::Units::distance::A, dot(), inverse(), and qmcplusplus::Units::force::N.
Referenced by MPC::init_f_G().
|
inline |
Definition at line 150 of file MPC.cpp.
References qmcplusplus::Units::distance::A, dot(), inverse(), and qmcplusplus::Units::force::N.
| embt qmcplusplus::fakeMainScalarSamples | ( | ) |
| embt qmcplusplus::fakeSomeOperatorEstimatorSamples | ( | c-> | rank() | ) |
| void qmcplusplus::fill_from_text | ( | int | num_opt_crowds, |
| FillData & | fd | ||
| ) |
Definition at line 147 of file test_QMCCostFunctionBatched.cpp.
References CHECK(), comm, OHMMS::Controller, LinearMethodTestSupport::costFn, FillData::derivRecords, FillData::energy_new, QMCCostFunctionBase::ENERGY_NEW, QMCCostFunctionBatched::fillOverlapHamiltonianMatrices(), LinearMethodTestSupport::getDerivRecords(), LinearMethodTestSupport::getHDerivRecords(), LinearMethodTestSupport::getRecordsOnNode(), LinearMethodTestSupport::getSumValue(), ham, FillData::ham_gold, FillData::HDerivRecords, qmcplusplus::Units::force::N, FillData::numParam, FillData::numSamples, FillData::ovlp_gold, FillData::reweight, QMCCostFunctionBase::REWEIGHT, LinearMethodTestSupport::set_samples_and_param(), FillData::sum_e_wgt, QMCCostFunctionBase::SUM_E_WGT, FillData::sum_esq_wgt, QMCCostFunctionBase::SUM_ESQ_WGT, FillData::sum_wgt, and QMCCostFunctionBase::SUM_WGT.
Referenced by TEST_CASE().
| void qmcplusplus::fillIdentityMatrix | ( | Matrix< T > & | m | ) |
Definition at line 26 of file createTestMatrix.h.
References qmcplusplus::Units::distance::m.
Referenced by TEMPLATE_TEST_CASE(), and TEST_CASE().
|
inline |
Definition at line 243 of file LatticeAnalyzer.h.
References found_shorter_base().
Referenced by DTD_BConds< T, 3, PPPG >::DTD_BConds(), and DTD_BConds< T, 3, PPPG+SOA_OFFSET >::DTD_BConds().
|
inline |
rotate the state after 3dfft
Definition at line 285 of file einspline_helper.hpp.
References atan2(), Array< T, D, ALLOC >::data(), real(), and sincos().
|
inline |
Definition at line 130 of file einspline_helper.hpp.
References compute_phase().
Referenced by OneSplineOrbData::fft_spline().
|
inline |
Definition at line 158 of file einspline_helper.hpp.
References compute_phase(), and Array< T, D, ALLOC >::data().
|
inline |
rotate the state after 3dfft
First, add the eikr phase factor. Then, rotate the phase of the orbitals so that neither the real part nor the imaginary part are very near zero. This sometimes happens in crystals with high symmetry at special k-points.
Definition at line 70 of file einspline_helper.hpp.
References Array< T, D, ALLOC >::data(), qmcplusplus::Units::time::s, sincos(), and sqrt().
Referenced by OneSplineOrbData::fft_spline().
|
inline |
Definition at line 153 of file SmallMatrixDetCalculator.h.
References n.
Referenced by CustomizedMatrixDet< 1 >::evaluate(), CustomizedMatrixDet< 2 >::evaluate(), CustomizedMatrixDet< 3 >::evaluate(), CustomizedMatrixDet< 4 >::evaluate(), and CustomizedMatrixDet< 5 >::evaluate().
|
inline |
Definition at line 96 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by SpinDensityNew::accumulate(), NESpaceGrid< REAL >::accumulate(), DTD_BConds< T, 3, PPPG >::apply_bc(), DTD_BConds< T, 3, PPNG >::apply_bc(), SpaceGrid::check_grid(), NESpaceGrid< REAL >::check_grid(), MagnetizationDensity::computeBin(), DTD_BConds< T, 3, PPPG+SOA_OFFSET >::computeDist(), DTD_BConds< T, 3, PPNG+SOA_OFFSET >::computeDist(), DTD_BConds< T, 3, PPPG+SOA_OFFSET >::computeDistances(), DTD_BConds< T, 3, PPNG+SOA_OFFSET >::computeDistances(), DTD_BConds< T, 3, PPPG+SOA_OFFSET >::computeDistancesOffload(), DTD_BConds< T, 3, PPNG+SOA_OFFSET >::computeDistancesOffload(), SplineR2R< ST >::convertPos(), NESpaceGrid< REAL >::copyToSoA(), ECPComponentBuilder::doBreakUp(), MPC::evalLR(), DensityEstimator::evaluate(), SpinDensity::evaluate(), SpaceGrid::evaluate(), qmcplusplus::ewaldref::ewaldEnergy(), KContainer::findApproxMMax(), CubicBsplineGrid< T, LINEAR_1DGRID, PBC_CONSTRAINTS >::getGridPoint(), CubicBsplineGrid< T, LINEAR_1DGRID, FIRSTDERIV_CONSTRAINTS >::getGridPoint(), getSplineBound(), SpaceGrid::initialize_rectilinear(), FnFloor::operator()(), parseGridInput(), qmcplusplus::simd::remainder(), OptimalGrid::ReverseMap(), OptimalGrid2::ReverseMap(), CenterGrid::ReverseMap(), LogGrid::ReverseMap(), ClusterGrid::ReverseMap(), kSpaceJastrow::setupGvecs(), OneDimCubicSplineLinearGrid< T >::splint(), CrystalLattice< ST, 3 >::toUnit_floor(), and EshdfFile::writeQboxElectrons().
|
inline |
Definition at line 1443 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 327 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by FnFmod::operator()().
|
inline |
Definition at line 577 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 827 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1053 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1254 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1674 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1900 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 2101 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
| qmcplusplus::for | ( | ) |
Referenced by SlaterDetBuilder::createMSDFast(), SkAllEstimator::evaluate(), ForceChiesaPBCAA::initBreakup(), StressPBC::initBreakup(), CoulombPBCAB::initBreakup(), CountingGaussian::makeClone(), WaveFunctionTester::runRatioTest(), WalkerControlMPI::swapWalkersSimple(), TEST_CASE(), BackflowTransformation::testDeriv(), DiracDeterminantWithBackflow::testGG(), DiracDeterminantWithBackflow::testGGG(), and BackflowTransformation::transformOnly().
|
inline |
Definition at line 87 of file ForEach.h.
References ForEach< Expr, FTag, CTag >::apply(), and qmcplusplus::Units::charge::e.
Referenced by evaluate().
|
inline |
Definition at line 208 of file LatticeAnalyzer.h.
Referenced by find_reduced_basis().
|
inline |
Definition at line 80 of file EinsplineSetBuilderCommon.cpp.
References IntPart().
Referenced by EinsplineSetBuilder::AnalyzeTwists2(), and EinsplineSetBuilder::TileIons().
| std::vector<double> qmcplusplus::gauss_random_vals | ( | size_test *3+(size_test *3) % 2+ | size_test | ) |
| void generateCuspInfo | ( | Matrix< CuspCorrectionParameters > & | info, |
| const ParticleSet & | targetPtcl, | ||
| const ParticleSet & | sourcePtcl, | ||
| const LCAOrbitalSet & | lcao, | ||
| const std::string & | id, | ||
| Communicate & | Comm | ||
| ) |
Definition at line 661 of file CuspCorrectionConstruction.cpp.
References abs(), SpeciesSet::addAttribute(), app_log(), broadcastCuspInfo(), LCAOrbitalSet::C, OneMolecularOrbital::changeOrbital(), Matrix< T, Alloc >::cols(), CuspCorrection::cparam, createGlobalTimer(), FairDivideLow(), SPOSet::getOrbitalSetSize(), ParticleSet::getSpeciesSet(), ParticleSet::GroupID, LCAOrbitalSet::isIdentity(), LCAOrbitalSet::isOMPoffload(), minimizeForRc(), LCAOrbitalSet::myBasisSet, OneMolecularOrbital::phi(), Communicate::rank(), Matrix< T, Alloc >::rows(), Communicate::size(), splitPhiEta(), timer_level_fine, and timer_level_medium.
Referenced by LCAOrbitalBuilder::createSPOSetFromXML().
|
static |
Definition at line 47 of file ParticleSet.cpp.
References PS_accept, PS_donePbyP, PS_dt_move, PS_loadWalker, PS_mw_copy, PS_newpos, and PS_update.
|
inline |
Create a random 3D rotation matrix using a random generator.
Definition at line 25 of file RandomRotationMatrix.h.
Referenced by NonLocalECPotential::evalIonDerivsImpl(), SOECPotential::evaluateImpl(), NonLocalECPotential::evaluateImpl(), NonLocalECPotential::evaluateOneBodyOpMatrix(), NonLocalECPotential::evaluateOneBodyOpMatrixForceDeriv(), SOECPotential::evaluateValueAndDerivatives(), NonLocalECPotential::evaluateValueAndDerivatives(), SOECPotential::mw_evaluateImpl(), and NonLocalECPotential::mw_evaluateImpl().
|
inline |
Create a random 3D rotation matrix from three random numbers.
Each input random number should be in the range [0,1). See the script Numerics/tests/derive_random_rotation.py for more information about the algorithm.
Definition at line 27 of file RotationMatrix3D.h.
References cos(), sin(), and sqrt().
|
inline |
Definition at line 96 of file ParticleAttrib.h.
Referenced by WalkerConfigurations::putConfigurations(), HDFWalkerInput_0_4::read_hdf5(), HDFWalkerInput_0_4::read_hdf5_scatter(), and HDFWalkerInput_0_4::read_phdf5().
| void get_gridinfo_from_posgrid | ( | const std::vector< PosType > & | posgridlist, |
| const IndexType & | axis, | ||
| RealType & | lx, | ||
| RealType & | rx, | ||
| RealType & | dx, | ||
| IndexType & | Nx | ||
| ) |
|
inline |
map C types to hdf5 native types bool is explicit removed due to the fact that it is implementation-dependant
Referenced by h5d_append(), h5d_check_type(), h5d_read(), and h5d_write().
|
inline |
Definition at line 102 of file ParticleAttrib.h.
Referenced by WalkerConfigurations::putConfigurations().
| std::string qmcplusplus::get_leaf_name | ( | const std::string & | stack_name | ) |
Definition at line 219 of file TimerManager.cpp.
Referenced by TimerManager< qmcplusplus::TimerType< CLOCK > >::output_timing(), and TimerManager< qmcplusplus::TimerType< CLOCK > >::print_stack().
| int qmcplusplus::get_level | ( | const std::string & | stack_name | ) |
| ParticleSet qmcplusplus::get_two_species_particleset | ( | const SimulationCell & | simulation_cell | ) |
Definition at line 59 of file test_J2_derivatives.cpp.
References SpeciesSet::addSpecies(), ParticleSet::addTable(), ParticleSet::create(), ParticleSet::getSpeciesSet(), ParticleSet::R, ParticleSet::resetGroups(), ParticleSet::setName(), and ParticleSet::update().
Referenced by TEST_CASE().
|
inline |
Definition at line 35 of file CUDAallocator.hpp.
References CUDAallocator_device_mem_allocated.
Referenced by print_mem().
| void getCurrentLocalEnergy | ( | const ValueVector & | pos, |
| RealType | Zeff, | ||
| RealType | Rc, | ||
| RealType | originalELatRc, | ||
| CuspCorrection & | cusp, | ||
| OneMolecularOrbital & | phiMO, | ||
| ValueVector & | ELcurr | ||
| ) |
Compute effective local energy at vector of points.
| pos | input vector of radial distances |
| Zeff | effective charge from getZeff |
| Rc | cutoff radius |
| originalELatRc | Local energy at the center from the uncorrected orbital |
| cusp | cusp correction parameters |
| phiMO | uncorrected orbital (S-orbitals on this center only) |
| ELcurr | output local energy at each distance in pos |
Definition at line 374 of file CuspCorrectionConstruction.cpp.
References CuspCorrection::d2pr(), CuspCorrection::dpr(), qmcplusplus::Units::charge::e, OneMolecularOrbital::phi_vgl(), phiBar(), and CuspCorrection::Rr().
Referenced by evaluateForPhi0Body().
|
inline |
free template function to read the (user) dimension and shape of the dataset.
The dimensions contributed by T is excluded.
| T | data type supported by h5_space_type |
| grp | group id |
| aname | name of the dataspace |
| sizes_out | sizes of each direction. For a scalar, sizes_out.size() == 0 |
For example, if the data on the file is Matrix<TinyVector<std::complex<double>, 3>> The dataset on the file has a rank of 2 (matrix) + 1 (TinyVector) + 1 (std::complex) + 0 (double) = 4 getDataShape<TinyVector<std::complex<double>, 3>> only returns the first 2 dimension getDataShape<std::complex<double>> only returns the first 3 dimension getDataShape<double> returns all the 4 dimension
Definition at line 45 of file hdf_wrapper_functions.h.
References rank.
|
inline |
Definition at line 24 of file DriftOperators.h.
Referenced by setScaledDrift().
|
inline |
Definition at line 34 of file DualAllocator.hpp.
References dual_device_mem_allocated.
| RealType getELchi2 | ( | const ValueVector & | ELcurr, |
| const ValueVector & | ELideal | ||
| ) |
Sum of squares difference between the current and ideal local energies This is the objective function to be minimized.
| Elcurr | current local energy |
| Elideal | ideal local energy |
Definition at line 432 of file CuspCorrectionConstruction.cpp.
Referenced by evaluateForPhi0Body().
| TimerManager< NewTimer > & getGlobalTimerManager | ( | ) |
Definition at line 44 of file TimerManager.cpp.
Referenced by createGlobalTimer(), main(), and QMCMain::QMCMain().
| void getIdealLocalEnergy | ( | const ValueVector & | pos, |
| RealType | Z, | ||
| RealType | Rc, | ||
| RealType | ELorigAtRc, | ||
| ValueVector & | ELideal | ||
| ) |
Ideal local energy at a vector of points.
| pos | input vector of radial distances |
| Z | nuclear charge |
| Rc | cutoff radius where the correction meets the actual orbital |
| ELorigAtRc | local energy at Rc. beta0 is adjusted to make energy continuous at Rc |
| ELideal | - output the ideal local energy at pos values |
Definition at line 317 of file CuspCorrectionConstruction.cpp.
References getOneIdealLocalEnergy().
Referenced by minimizeForPhiAtZero().
| T* qmcplusplus::getOffloadDevicePtr | ( | T * | host_ptr | ) |
Definition at line 38 of file OMPallocator.hpp.
Referenced by OMPallocator< Value >::allocate(), and OMPallocator< Value >::deallocate().
|
inline |
Definition at line 35 of file OMPallocator.hpp.
References OMPallocator_device_mem_allocated.
Referenced by print_mem().
Ideal local energy at one point.
| r | input radial distance |
| Z | nuclear charge |
| beta0 | adjustable parameter to make energy continuous at Rc |
Definition at line 303 of file CuspCorrectionConstruction.cpp.
Referenced by getIdealLocalEnergy().
| RealType getOriginalLocalEnergy | ( | const ValueVector & | pos, |
| RealType | Zeff, | ||
| RealType | Rc, | ||
| OneMolecularOrbital & | phiMO, | ||
| ValueVector & | Elorig | ||
| ) |
Local energy from uncorrected orbital.
| pos | input vector of radial distances |
| Zeff | nuclear charge |
| Rc | cutoff radius |
| phiMO | uncorrected orbital (S-orbitals on this center only) |
| ELorig | output local energy at each distance in pos |
Return is value of local energy at zero. This is the value needed for subsequent computations. The routine can be called with an empty vector of positions to get just this value.
Definition at line 408 of file CuspCorrectionConstruction.cpp.
References OneMolecularOrbital::phi_vgl().
Referenced by minimizeForPhiAtZero().
|
inline |
evaluate the determinant ratio with a column substitution
Definition at line 140 of file determinant_operators.h.
References BLAS::dot(), and qmcplusplus::Units::distance::m.
|
inline |
|
inline |
evaluate the ratio with a column substitution and multiple row substitutions
| refinv | reference inverse(m,m) |
| tcm | new column whose size > m |
| ratios | ratios of the row substitutions with the column substitution |
| m | dimension |
| colchanged | ratio with a column substitution of refinv |
| r_replaced | row index for the excitations in ind |
| ind | indexes for the excited states (rows) |
Definition at line 164 of file determinant_operators.h.
References BLAS::dot(), and qmcplusplus::Units::distance::m.
|
inline |
Definition at line 106 of file determinant_operators.h.
References BLAS::gemv(), and qmcplusplus::Units::distance::m.
|
inline |
Definition at line 116 of file determinant_operators.h.
References BLAS::dot(), and qmcplusplus::Units::distance::m.
|
inline |
Definition at line 127 of file determinant_operators.h.
References BLAS::dot(), and qmcplusplus::Units::distance::m.
|
inline |
evaluate a drift with a real force
| tau | timestep |
| qf | quantum force |
| drift |
Definition at line 36 of file DriftOperators.h.
References convertToReal(), dot(), and sqrt().
Referenced by DMCUpdatePbyPL2::advanceWalker(), and assignDrift().
|
inline |
evaluate a drift with a real force with rescaling for an L2 potential
| tau | timestep |
| qf | quantum force |
| drift |
Definition at line 52 of file DriftOperators.h.
References convertToReal(), dot(), and sqrt().
Referenced by DMCUpdatePbyPL2::advanceWalker().
|
inline |
break x into the integer part and residual part and apply bounds
| x | input coordinate |
| nmax | input upper bound of the integer part |
| ind | output integer part |
| dx | output fractional part |
x in the range of [0, nmax+1) will be split correctly. x < 0, ind = 0, dx = 0 x >= nmax+1, ind = nmax, dx = 1 - epsilon
Attention: nmax is not the number grid points but the maximum allowed grid index For example, ng is the number of grid point. the actual grid points indices are 0, 1, ..., ng - 1. In a periodic/anti periodic spline, set nmax = ng - 1 In a natural boundary spline, set nmax = ng - 2 because the end point should be excluded and the last grid point has an index ng - 2.
Definition at line 37 of file SplineBound.hpp.
References floor().
Referenced by BsplineFunctor< REAL >::evaluate(), BsplineFunctor< REAL >::evaluate_impl(), BsplineFunctor< REAL >::evaluateDerivatives(), BsplineFunctor< REAL >::evaluateV(), BsplineFunctor< REAL >::evaluateVGL(), and test_spline_bounds().
| QMCTraits::RealType qmcplusplus::getSplinedSOPot | ( | SOECPComponent * | so_comp, |
| int | l, | ||
| double | r | ||
| ) |
Definition at line 44 of file test_ecp.cpp.
References SOECPComponent::sopp_m_.
Referenced by TEST_CASE().
| void getStats | ( | const std::vector< RealType > & | vals, |
| RealType & | avg, | ||
| RealType & | err, | ||
| int | start = 0 |
||
| ) |
Simpleaverage and error estimate.
input array of values to be averaged. returns avg and err. Can pass a start index to average from "start" to the end of the array
Definition at line 31 of file FSUtilities.cpp.
Referenced by QMCFiniteSize::calcLeadingOrderCorrections(), QMCFiniteSize::calcPotentialCorrection(), estimateEquilibration(), and SkParserHDF5::parse().
| sycl::queue & getSYCLDefaultDeviceDefaultQueue | ( | ) |
return a reference to the per-device default queue
Definition at line 18 of file SYCLruntime.cpp.
References SYCLDeviceManager::getDefaultDeviceDefaultQueue().
Referenced by SYCLSharedAllocator< T, ALIGN >::allocate(), SYCLAllocator< std::int64_t >::allocate(), SYCLHostAllocator< int >::allocate(), SYCLAllocator< std::int64_t >::copyDeviceToDevice(), SYCLAllocator< std::int64_t >::copyFromDevice(), SYCLAllocator< std::int64_t >::copyToDevice(), createSYCLInOrderQueueOnDefaultDevice(), createSYCLQueueOnDefaultDevice(), SYCLSharedAllocator< T, ALIGN >::deallocate(), SYCLAllocator< std::int64_t >::deallocate(), SYCLHostAllocator< int >::deallocate(), getSYCLdeviceFreeMem(), TEST_CASE(), and test_inverse().
| size_t getSYCLdeviceFreeMem | ( | ) |
query free memory on the default device
Definition at line 31 of file SYCLruntime.cpp.
References getSYCLDefaultDeviceDefaultQueue().
Referenced by print_mem().
|
inline |
Definition at line 37 of file SYCLallocator.hpp.
References SYCLallocator_device_mem_allocated.
Referenced by print_mem().
just set the values to test the electron weights
Definition at line 309 of file test_bare_kinetic.cpp.
References CHECK().
Referenced by TEST_CASE().
|
inline |
evaluate a drift with a real force and no rescaling
| tau | timestep |
| qf | quantum force |
| drift |
Definition at line 70 of file DriftOperators.h.
References convertToReal().
Effective nuclear charge to keep effective local energy finite at zero.
| Z | nuclear charge |
| etaAtZero | value of non-S orbitals at this center |
| phiBarAtZero | value of corrected orbital at zero |
Definition at line 362 of file CuspCorrectionConstruction.cpp.
Referenced by evaluateForPhi0Body(), and minimizeForPhiAtZero().
|
inline |
Definition at line 399 of file hdf_wrapper_functions.h.
References dims, and get_h5_datatype().
Referenced by ObservableHelper::write(), TraceBuffer< TraceInt >::write_hdf(), and WalkerLogBuffer< WLog::Real >::writeHDF().
|
inline |
Definition at line 136 of file hdf_wrapper_functions.h.
Referenced by h5data_proxy< T >::check_existence().
|
inline |
Definition at line 151 of file hdf_wrapper_functions.h.
References get_h5_datatype().
|
inline |
return true, if successful
Definition at line 123 of file hdf_wrapper_functions.h.
References get_h5_datatype().
Referenced by h5data_proxy< Vector< T > >::read(), h5data_proxy< std::vector< T > >::read(), h5data_proxy< T >::read(), h5data_proxy< boost::multi::array< T, 1, Alloc > >::read(), h5data_proxy< bool >::read(), h5data_proxy< Matrix< T > >::read(), h5data_proxy< accumulator_set< T > >::read(), h5data_proxy< boost::multi::array< T, 2, Alloc > >::read(), h5data_proxy< std::vector< bool > >::read(), h5data_proxy< Array< T, D > >::read(), h5data_proxy< boost::multi::array_ref< T, 1, Ptr > >::read(), h5data_proxy< double_hyperslab_proxy< CT, MAXDIM > >::read(), h5data_proxy< boost::multi::array_ref< T, 2, Ptr > >::read(), and h5data_proxy< hyperslab_proxy< CT, RANK > >::read().
| bool qmcplusplus::h5d_read | ( | hid_t | grp, |
| const std::string & | aname, | ||
| hsize_t | ndims, | ||
| const hsize_t * | gcounts, | ||
| const hsize_t * | counts, | ||
| const hsize_t * | offsets, | ||
| T * | first, | ||
| hid_t | xfer_plist | ||
| ) |
return true, if successful
Definition at line 210 of file hdf_wrapper_functions.h.
References get_h5_datatype().
| bool qmcplusplus::h5d_read | ( | hid_t | grp, |
| const std::string & | aname, | ||
| hsize_t | ndims, | ||
| const hsize_t * | gcounts, | ||
| const hsize_t * | counts, | ||
| const hsize_t * | offsets, | ||
| hsize_t | mem_ndims, | ||
| const hsize_t * | mem_gcounts, | ||
| const hsize_t * | mem_counts, | ||
| const hsize_t * | mem_offsets, | ||
| T * | first, | ||
| hid_t | xfer_plist | ||
| ) |
return true, if successful
Definition at line 302 of file hdf_wrapper_functions.h.
References get_h5_datatype().
|
inline |
Definition at line 175 of file hdf_wrapper_functions.h.
References dims, and get_h5_datatype().
Referenced by h5data_proxy< Vector< T > >::write(), h5data_proxy< std::vector< T > >::write(), h5data_proxy< T >::write(), h5data_proxy< boost::multi::array< T, 1, Alloc > >::write(), h5data_proxy< accumulator_set< T > >::write(), h5data_proxy< Matrix< T > >::write(), h5data_proxy< bool >::write(), h5data_proxy< boost::multi::array< T, 2, Alloc > >::write(), h5data_proxy< std::vector< bool > >::write(), h5data_proxy< Array< T, D > >::write(), h5data_proxy< boost::multi::array_ref< T, 1, Ptr > >::write(), h5data_proxy< double_hyperslab_proxy< CT, MAXDIM > >::write(), h5data_proxy< boost::multi::array_ref< T, 2, Ptr > >::write(), and h5data_proxy< hyperslab_proxy< CT, RANK > >::write().
|
inline |
Definition at line 252 of file hdf_wrapper_functions.h.
References get_h5_datatype().
|
inline |
Definition at line 342 of file hdf_wrapper_functions.h.
References get_h5_datatype().
| constexpr bool qmcplusplus::has | ( | const R & | this_one | ) |
Definition at line 23 of file variant_help.hpp.
Referenced by MagnetizationDensityInput::MagnetizationDensityInputSection::checkParticularValidity(), and CustomTestInput::setFromStreamCustom().
| if | ( | c-> | rank() = = 0 | ) |
Definition at line 76 of file test_manager_mpi.cpp.
References CHECK(), EstimatorManagerNewTest::em, embt, EstimatorManagerNew::get_AverageCache(), and num_ranks.
Referenced by accum_sample(), ci_configuration2::calculateExcitations(), WalkerLogCollector::collect(), EnergyDensityEstimator::evaluate(), VirtualParticleSet::makeMoves(), EnergyDensityEstimator::put(), eeI_JastrowBuilder::putkids(), QMCDriverFactory::readSection(), SPOSetScanner::scan_path(), and EnergyDensityEstimator::set_ptcl().
|
inline |
imaginary part of a scalar. Cannot be replaced by std::imag due to AFQMC specific needs.
Definition at line 91 of file complex_help.hpp.
Referenced by RotatedSPOs::buildOptVariables(), TrialWaveFunction::calcRatioGrad(), TrialWaveFunction::calcRatioGradWithSpin(), calcSOECP(), Quadrature3D< T >::CheckQuadratureRule(), WalkerLogCollector::collect(), WalkerLogBuffer< WLog::Real >::collect(), LogToValue< std::complex< T > >::convert(), kSpaceJastrow::evalGrad(), DensityMatrices1B::evaluate_loop(), DensityMatrices1B::evaluate_matrix(), TrialWaveFunction::evaluateDeltaLog(), kSpaceJastrow::evaluateDerivatives(), TrialWaveFunction::evaluateGL(), kSpaceJastrow::evaluateLog(), TrialWaveFunction::evaluateLog(), OneBodyDensityMatrices::evaluateMatrix(), RotatedSPOs::exponentiate_antisym_matrix(), ComplexExpFitClass< M >::Fit(), ComplexExpFitClass< M >::FitCusp(), TrialWaveFunction::getPhases(), RotatedSPOs::log_antisym_matrix(), main(), TrialWaveFunction::mw_accept_rejectMove(), TrialWaveFunction::mw_evaluateGL(), TrialWaveFunction::mw_evaluateLog(), OrthogExcluding(), kSpaceJastrow::ratioGrad(), WaveFunctionTester::runZeroVarianceTest(), test_C_diamond(), TEST_CASE(), test_diamond_2x1x1_xml_input(), test_gemm(), test_LCAO_DiamondC_2x1x1_cplx(), test_LCAO_DiamondC_2x1x1_real(), test_one_gemm(), TrialWaveFunction::updateBuffer(), and OrbitalImages::write_orbital_xsf().
|
inline |
Definition at line 92 of file complex_help.hpp.
|
inline |
Definition at line 93 of file complex_help.hpp.
|
inline |
Definition at line 94 of file complex_help.hpp.
|
inline |
Definition at line 41 of file kSpaceJastrow.cpp.
Referenced by kSpaceJastrow::setupGvecs().
|
inline |
Definition at line 55 of file kSpaceJastrow.cpp.
| std::vector<int, CUDAHostAllocator<int> > qmcplusplus::infos | ( | 8 | , |
| 1. | 0 | ||
| ) |
Referenced by TEST_CASE().
| void qmcplusplus::init_string_output | ( | ) |
Definition at line 33 of file test_output_manager.cpp.
References app_out, debug_out, err_out, infoDebug, infoError, infoLog, infoSummary, reset_string_output(), InfoStream::setStream(), and summary_out.
Referenced by TEST_CASE().
| template bool qmcplusplus::InputSection::setIfInInput< qmcplusplus::MagnetizationDensityInput::Integrator > | ( | qmcplusplus::MagnetizationDensityInput::Integrator & | var, |
| const std::string & | tag | ||
| ) |
| template bool qmcplusplus::InputSection::setIfInInput< qmcplusplus::OneBodyDensityMatricesInput::Integrator > | ( | qmcplusplus::OneBodyDensityMatricesInput::Integrator & | var, |
| const std::string & | tag | ||
| ) |
| template bool qmcplusplus::InputSection::setIfInInput< ReferencePointsInput::Coord > | ( | ReferencePointsInput::Coord & | var, |
| const std::string & | tag | ||
| ) |
|
inline |
Definition at line 119 of file string_utils.h.
Referenced by SpaceGrid::registerCollectables(), NESpaceGrid< REAL >::registerGrid(), SHOSetBuilder::report(), SHOSet::report(), and SHOSet::test_derivatives().
|
inline |
Definition at line 74 of file EinsplineSetBuilderCommon.cpp.
References qmcplusplus::Units::charge::e.
Referenced by EinsplineSetBuilder::AnalyzeTwists2(), and FracPart().
Definition at line 879 of file TensorOps.h.
Referenced by DTD_BConds< T, 3, PPPG >::DTD_BConds(), DTD_BConds< T, 3, PPPG+SOA_OFFSET >::DTD_BConds(), MultiDiracDeterminant::evaluateDetsAndGradsForPtclMove(), MultiDiracDeterminant::evaluateDetsAndGradsForPtclMoveWithSpin(), MultiDiracDeterminant::evaluateDetsForPtclMove(), qmcplusplus::ewaldref::ewaldEnergy(), qmcplusplus::ewaldref::ewaldSum(), extrap(), SpaceGrid::initialize_rectilinear(), qmcplusplus::ewaldref::madelungSum(), TinyVector< Real, OHMMS_DIM >::operator-(), NESpaceGrid< REAL >::processAxis(), EinsplineSetBuilder::ReadOrbitalInfo_ESHDF(), and CrystalLattice< ST, 3 >::reset().
Definition at line 888 of file TensorOps.h.
Definition at line 907 of file TensorOps.h.
References det().
|
inline |
Definition at line 269 of file DeterminantOperators.h.
References Matrix< T, Alloc >::data(), Vector< T, Alloc >::data(), det_col_update(), and Matrix< T, Alloc >::rows().
Referenced by AGPDeterminant::acceptMove(), MultiDiracDeterminant::evaluateDetsAndGradsForPtclMove(), MultiDiracDeterminant::evaluateDetsAndGradsForPtclMoveWithSpin(), MultiDiracDeterminant::evaluateDetsForPtclMove(), MultiDiracDeterminant::evaluateForWalkerMove(), MultiDiracDeterminant::evaluateForWalkerMoveWithSpin(), MultiDiracDeterminant::evaluateGrads(), MultiDiracDeterminant::evaluateGradsWithSpin(), and AGPDeterminant::ratioDown().
|
inline |
update a inverse matrix by a row substitution
| Minv | in/out inverse matrix |
| newrow | row vector |
| rvec | workspace |
| rvecinv | workspace |
| rowchanged | row index to be replaced |
| c_ratio | determinant-ratio with the row replacement |
Definition at line 247 of file DeterminantOperators.h.
References Matrix< T, Alloc >::cols(), Matrix< T, Alloc >::data(), Vector< T, Alloc >::data(), and det_row_update().
Referenced by AGPDeterminant::acceptMove(), and AGPDeterminant::ratioUp().
|
inline |
inverse a matrix
| x | starting address of an n-by-m matrix |
| n | rows |
| m | cols |
| work | workspace array |
| pivot | integer pivot array |
Definition at line 118 of file DeterminantOperators.h.
References InvertLU(), LUFactorization(), qmcplusplus::Units::distance::m, and n.
Referenced by RotatedSPOs::evaluateDerivatives(), RotatedSPOs::evaluateDerivativesWF(), RotatedSPOs::evaluateDerivRatios(), and Invert().
|
inline |
inverse a matrix
| x | starting address of an n-by-m matrix |
| n | rows |
| m | cols |
Workspaces are handled internally.
Definition at line 164 of file DeterminantOperators.h.
References Invert(), qmcplusplus::Units::distance::m, and n.
|
inline |
invert a matrix
| M | a matrix to be inverted |
| getdet | bool, if true, calculate the determinant |
Definition at line 187 of file DeterminantOperators.h.
References InvertLU(), LUFactorization(), n, and sign().
Referenced by ExpFitClass< M >::Fit(), ExpFitClass< M >::FitCusp(), ForceCeperley::InitMatrix(), ForceChiesaPBCAA::InitMatrix(), ForceBase::initVarReduction(), TWFFastDerivWrapper::invertMatrices(), LinearFit(), QMCFixedSampleLinearOptimizeBatched::previous_linear_methods_run(), QMCFixedSampleLinearOptimize::run(), Eigensolver::solveGeneralizedEigenvalues_Inv(), and QMCFixedSampleLinearOptimizeBatched::solveShiftsWithoutLMYEngine().
|
inline |
Inversion of a double matrix after LU factorization.
Definition at line 65 of file DeterminantOperators.h.
Referenced by Invert(), invert_matrix(), and InvertWithLog().
|
inline |
Inversion of a float matrix after LU factorization.
Definition at line 72 of file DeterminantOperators.h.
|
inline |
Inversion of a std::complex<double> matrix after LU factorization.
Definition at line 84 of file DeterminantOperators.h.
|
inline |
Inversion of a complex<float> matrix after LU factorization.
Definition at line 96 of file DeterminantOperators.h.
|
inline |
Definition at line 172 of file DeterminantOperators.h.
References InvertLU(), log(), LUFactorization(), qmcplusplus::Units::distance::m, and n.
Referenced by DiracDeterminantWithBackflow::dummyEvalLi(), DiracDeterminantWithBackflow::evaluateDerivatives(), MultiDiracDeterminant::evaluateForWalkerMove(), MultiDiracDeterminant::evaluateForWalkerMoveWithSpin(), DiracDeterminantWithBackflow::evaluateLog(), AGPDeterminant::evaluateLogAndStore(), DiracDeterminantWithBackflow::ratio(), DiracDeterminantWithBackflow::ratioGrad(), RotatedSPOs::table_method_eval(), RotatedSPOs::table_method_evalWF(), DiracDeterminantWithBackflow::testDerivFjj(), and DiracDeterminantWithBackflow::testL().
|
inline |
Definition at line 116 of file LCAOrbitalBuilder.cpp.
Referenced by RadialJastrowBuilder::buildComponent(), SpaceGridInput::SpaceGridInputSection::checkParticularValidity(), checkShapeConsistency(), LCAOrbitalBuilder::loadBasisSetFromXML(), and TEST_CASE().
| bool isfinite | ( | float | ) |
return true if the value is finite.
std::isfinite can be affected by compiler the -ffast-math option and return true constantly. The customized qmcplusplus::isnan should be always effective. This requires its definition compiled without -ffast-math.
Definition at line 21 of file math.cpp.
References isfinite().
Referenced by DMCBatched::advanceWalkers().
| bool isfinite | ( | double | a | ) |
| bool isinf | ( | float | ) |
return true if the value is Inf.
std::isinf can be affected by compiler the -ffast-math option and return true constantly. The customized qmcplusplus::isnan should be always effective. This requires its definition compiled without -ffast-math.
Definition at line 24 of file math.cpp.
References isinf().
Referenced by DiracDeterminantBase::checkG().
| bool isnan | ( | float | ) |
return true if the value is NaN.
std::isnan can be affected by compiler the -ffast-math option and return true constantly. The customized qmcplusplus::isnan should be always effective. This requires its definition compiled without -ffast-math.
Definition at line 18 of file math.cpp.
Referenced by DiracDeterminantBase::checkG(), NaNguard::checkOneParticleGradients(), NaNguard::checkOneParticleRatio(), HamiltonianRef::evaluate(), syclSolverInverter< T_FP >::invert_transpose(), cuSolverInverter< T_FP >::invert_transpose(), rocSolverInverter< T_FP >::invert_transpose(), QMCFixedSampleLinearOptimizeBatched::one_shift_run(), test_isnan(), QMCHamiltonian::updateComponent(), and DescentEngine::updateParameters().
|
inline |
Definition at line 77 of file math.hpp.
Referenced by accumulator_set< FullPrecReal >::bad(), and accumulator_set< FullPrecReal >::variance().
|
inline |
compute real(laplacian)
Definition at line 27 of file BareKineticHelper.h.
References dot().
Referenced by SpeciesKineticEnergy::evaluate(), BareKineticEnergy::evaluate(), BareKineticEnergy::evaluate_orig(), and BareKineticEnergy::evaluateWithIonDerivs().
|
inline |
specialization of laplacian with complex g & l
Definition at line 35 of file BareKineticHelper.h.
References OTCDot< T1, T2, D >::apply().
|
inline |
Definition at line 305 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by GKIntegration< F, GKRule >::Integrate(), and FnLdexp::operator()().
|
inline |
Definition at line 555 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 805 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1035 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1236 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1652 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1882 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 2083 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 25 of file Ylm.h.
References sqrt().
Referenced by LegendrePlm(), and TEST_CASE().
|
inline |
Definition at line 45 of file Ylm.h.
References abs(), LegendrePll(), qmcplusplus::Units::distance::m, and sign().
Referenced by Ylm().
|
inline |
Definition at line 28 of file LinearFit.h.
References qmcplusplus::Units::distance::A, invert_matrix(), and qmcplusplus::Units::force::N.
Referenced by BsplineFunctor< REAL >::initialize(), BackflowBuilder::makeShortRange_twoBody(), and BsplineFunctor< REAL >::put().
|
inline |
Definition at line 104 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by assignGaussRand(), AtomicOrbitals< ST >::AtomicOrbitals(), SimpleFixedNodeBranch::branch(), WaveFunctionTester::checkGradients(), computeLogDet(), computeLogDet_sycl(), convertValueToLog(), RPABFeeBreakup< T >::Dlindhard(), RPABFeeBreakup< T >::Dlindhardp(), ExampleHeComponent::evaluateLog(), evalX(), ExpFitClass< M >::Fit(), ExpFitClass< M >::FitCusp(), RunTimeControl< CLOCK >::generateProgressMessage(), RunTimeControl< CLOCK >::generateStopMessage(), LogGridLight< T >::getCL(), OptimalGrid::Init(), OptimalGrid2::Init(), CenterGrid::Init(), OptimalGrid::InitRatio(), InvertWithLog(), LogGridLight< T >::locate(), LogGrid< T, CT >::locate(), LogGridZero< T, CT >::locate(), RotatedSPOs::log_antisym_matrix(), OneBodyDensityMatrices::OneBodyDensityMatrices(), FnLog::operator()(), print_mem(), SimpleFixedNodeBranch::reset(), OptimalGrid::ReverseMap(), LogGrid::ReverseMap(), WaveFunctionTester::runGradSourceTest(), WaveFunctionTester::runZeroVarianceTest(), LogGridLight< T >::set(), LogGrid< T, CT >::set(), DensityMatrices1B::set_state(), EwaldHandlerQuasi2D::slabLogf(), TEST_CASE(), SHOSetBuilder::update_basis_states(), CSUpdateBase::updateNorms(), and SFNBranch::updateParamAfterPopControl().
|
inline |
Definition at line 1451 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 112 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by FnLog10::operator()().
|
inline |
Definition at line 1459 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
| std::vector<StdComp, CUDAHostAllocator<StdComp> > qmcplusplus::log_values | ( | batch_size | ) |
Referenced by QMCDriverNew::checkLogAndGL(), DiracMatrixComputeOMPTarget< VALUE_FP >::computeInvertAndLog(), DiracMatrixComputeCUDA< VALUE_FP >::invert_transpose(), DiracMatrixComputeCUDA< VALUE_FP >::mw_computeInvertAndLog(), DiracMatrixComputeCUDA< VALUE_FP >::mw_computeInvertAndLog_stride(), DiracMatrixComputeOMPTarget< VALUE_FP >::mw_invertTranspose(), DiracMatrixComputeCUDA< VALUE_FP >::mw_invertTranspose(), TEST_CASE(), and testTrialWaveFunction_diamondC_2x1x1().
|
inline |
helper function to determine the lower bound of a domain (need to move up)
Definition at line 201 of file InitMolecularSystem.cpp.
References omptarget::min().
Referenced by MultiQuinticSpline1D< T >::batched_evaluate(), MultiQuinticSpline1D< T >::batched_evaluateVGL(), InitMolecularSystem::initWithVolume(), EinsplineSetBuilder::ReadGvectors_ESHDF(), CollectablesEstimator::registerObservables(), LocalEnergyEstimator::registerObservables(), and unpack4fftw().
| std::string lowerCase | ( | const std::string_view | s | ) |
++17
take string_view (or something that can be implicitly converted to one) and return lcase string. Don't call on anything but ASCII strings.
According to en.cppreference.com std::tolower is only defined for usigned char and EOF. the std::tolower conversion is based on the c locale which to makes it unclear on what might happen to char > 127.
Definition at line 22 of file ModernStringUtils.cpp.
References qmcplusplus::Units::time::s.
Referenced by ParameterSet::add(), RadialJastrowBuilder::buildComponent(), createDriftModifier(), QMCGaussianParserBase::createHamiltonian(), RadialJastrowBuilder::createJ1(), SPOSetBuilderFactory::createSPOSetBuilder(), getNodeName(), LocalEnergyInput::LocalEnergyInput(), InputSection::lookupAnyEnum(), ProjectData::lookupDriverVersion(), QMCFixedSampleLinearOptimize::processOptXML(), LatticeParser::put(), ParameterSet::put(), QMCCostFunctionBase::put(), RadialOrbitalSetBuilder< SoaAtomicBasisSet< MultiFunctorAdapter< FN >, SH > >::put(), OhmmsParameter< bool >::put(), ShortRangeCuspFunctor< T >::put(), TraceManager::put(), InputSection::readAttributes(), QMCDriverInput::readXML(), EstimatorManagerInput::readXML(), InputSection::readXML(), ShortRangeCuspFunctor< T >::set_variable_from_xml(), ParameterSet::setValue(), and TEST_CASE().
| testing::MatrixAccessor<double> qmcplusplus::lu2_mat | ( | lu2. | data(), |
| 4 | , | ||
| 4 | |||
| ) |
| testing::MatrixAccessor<double> qmcplusplus::lu_mat | ( | lu. | data(), |
| 4 | , | ||
| 4 | |||
| ) |
Referenced by TEST_CASE().
|
inline |
LU factorization of double.
Definition at line 37 of file DeterminantOperators.h.
References dgetrf(), qmcplusplus::Units::distance::m, and n.
Referenced by Determinant(), Invert(), invert_matrix(), and InvertWithLog().
|
inline |
LU factorization of float.
Definition at line 44 of file DeterminantOperators.h.
References qmcplusplus::Units::distance::m, n, and sgetrf().
|
inline |
LU factorization of std::complex<double>
Definition at line 51 of file DeterminantOperators.h.
References qmcplusplus::Units::distance::m, n, and zgetrf().
|
inline |
LU factorization of complex<float>
Definition at line 58 of file DeterminantOperators.h.
References cgetrf(), qmcplusplus::Units::distance::m, and n.
| std::vector<StdComp*, CUDAHostAllocator<StdComp*> > qmcplusplus::lus | ( | batch_size | ) |
| testing::MatrixAccessor<double> qmcplusplus::M2_mat | ( | M2_vec. | data(), |
| 4 | , | ||
| 4 | |||
| ) |
| testing::MatrixAccessor<double> qmcplusplus::M_mat | ( | M_vec. | data(), |
| 4 | , | ||
| 4 | |||
| ) |
Referenced by TEST_CASE().
|
inline |
Definition at line 54 of file BsplineReader.cpp.
Referenced by BsplineReader::create_spline_set().
|
inline |
Definition at line 38 of file BsplineReader.cpp.
Referenced by BsplineReader::initialize_spo2band().
| std::any qmcplusplus::makeAnotherInput | ( | xmlNodePtr | cur, |
| std::string & | value_label | ||
| ) |
Definition at line 551 of file test_InputSection.cpp.
References AnotherInput::get_name().
Referenced by DelegatingInput::DelegatingInputSection::DelegatingInputSection().
|
inline |
Definition at line 25 of file RandomSeqGeneratorGlobal.h.
References assignGaussRand(), and Random.
Referenced by InitMolecularSystem::initAtom(), ParticleSet::randomizeFromSource(), WaveFunctionTester::run(), WaveFunctionTester::runBasicTest(), WaveFunctionTester::runRatioTest(), WaveFunctionTester::runRatioTest2(), WaveFunctionTester::runRatioV(), and WaveFunctionTester::WaveFunctionTester().
|
inline |
specialized functions: stick to overloading
Definition at line 32 of file RandomSeqGeneratorGlobal.h.
References assignGaussRand(), and Random.
|
inline |
Definition at line 38 of file RandomSeqGeneratorGlobal.h.
References assignGaussRand(), and Random.
|
inline |
Definition at line 44 of file RandomSeqGeneratorGlobal.h.
References assignGaussRand(), Random, and Vector< T, Alloc >::size().
|
inline |
Definition at line 71 of file RandomSeqGenerator.h.
References assignGaussRand().
Referenced by SOVMCUpdateAll::advanceWalker(), DMCUpdateAllWithRejection::advanceWalker(), SODMCUpdatePbyPWithRejectionFast::advanceWalker(), VMCUpdateAll::advanceWalker(), DMCUpdatePbyPWithRejectionFast::advanceWalker(), SOVMCUpdatePbyP::advanceWalker(), VMCUpdatePbyP::advanceWalker(), DMCUpdatePbyPL2::advanceWalker(), CSVMCUpdatePbyP::advanceWalker(), CSVMCUpdateAll::advanceWalker(), DMCUpdateAllWithKill::advanceWalker(), CSVMCUpdateAllWithDrift::advanceWalker(), VMCBatched::advanceWalkers(), DMCBatched::advanceWalkers(), RMCUpdatePbyPWithDrift::advanceWalkersRMC(), RMCUpdateAllWithDrift::advanceWalkersRMC(), RMCUpdatePbyPWithDrift::advanceWalkersVMC(), RMCUpdateAllWithDrift::advanceWalkersVMC(), QMCMain::executeCMCSection(), makeGaussRandomWithEngine(), and TEST_CASE().
|
inline |
Definition at line 77 of file RandomSeqGenerator.h.
References assignGaussRand().
|
inline |
Definition at line 83 of file RandomSeqGenerator.h.
References assignGaussRand().
|
inline |
Definition at line 91 of file RandomSeqGenerator.h.
References assignGaussRand(), and Vector< T, Alloc >::size().
|
inline |
Definition at line 97 of file RandomSeqGenerator.h.
References makeGaussRandomWithEngine(), and POS_SPIN.
| qmcplusplus::makeGaussRandomWithEngine | ( | gauss_random_vals | , |
| rng | |||
| ) |
| qmcplusplus::makeGaussRandomWithEngine | ( | mc_coords_rs | , |
| rng | |||
| ) |
| qmcplusplus::makeGaussRandomWithEngine | ( | mc_coords_rsspins | , |
| rng | |||
| ) |
| PSetsAndRefList qmcplusplus::makePsets | ( | ) |
Definition at line 87 of file test_ReferencePoints.cpp.
References comm, OHMMS::Controller, lattice, MinimalParticlePool::make_diamondC_1x1x1(), qmcplusplus::testing::makeTestLattice(), particle_pool, and pset.
Referenced by TEST_CASE().
| std::any makeReferencePointsInput | ( | xmlNodePtr | , |
| std::string & | value_label | ||
| ) |
factory function used by InputSection to make reference points Input
| [out] | value_label | key value in delegating InputSection for storing the constructed Input from processed node. |
Definition at line 83 of file ReferencePointsInput.cpp.
Referenced by TEST_CASE().
|
static |
}@
helper function to take vector of class A to refvector of any valid reference type for A
intended usage looks like this std::vector<DerivedType> vdt auto refvecbase = makeRefVector<BaseType>(vdt) or if you just want a refvector of type vdt auto refvec = makeRefVector<decltype(vdt)::value_type>(vdt)
godbolt.org indicates at least with clang 11&12 we get RVO here. auto ref_whatevers = makeRefVector<ValidTypeForReference>(whatevers); makes no extra copy.
Definition at line 54 of file template_types.hpp.
| std::any makeSpaceGridInput | ( | xmlNodePtr | , |
| std::string & | value_label | ||
| ) |
factory function for a SpaceGridInput
| [in] | input | node for SpaceGridInput |
| [out] | value | label returned to caller |
Definition at line 143 of file SpaceGridInput.cpp.
Referenced by TEST_CASE().
|
inline |
Definition at line 62 of file RandomSeqGeneratorGlobal.h.
References Random, and sqrt().
Referenced by InitMolecularSystem::initMolecule().
|
inline |
Definition at line 86 of file RandomSeqGeneratorGlobal.h.
References Random, and sqrt().
| ReferencePointsInput qmcplusplus::makeTestRPI | ( | ) |
Definition at line 70 of file test_ReferencePoints.cpp.
References doc, Libxml2Document::getRoot(), node, okay, and Libxml2Document::parseFromString().
Referenced by TEST_CASE().
|
inline |
Definition at line 50 of file RandomSeqGeneratorGlobal.h.
References assignUniformRand(), and Random.
Referenced by InitMolecularSystem::initWithVolume(), InitMolecularSystem::put(), and XMLParticleParser::readXML().
|
inline |
Definition at line 56 of file RandomSeqGeneratorGlobal.h.
References assignUniformRand(), Random, and Vector< T, Alloc >::size().
| RealType minimizeForPhiAtZero | ( | CuspCorrection & | cusp, |
| OneMolecularOrbital & | phiMO, | ||
| RealType | Z, | ||
| RealType | eta0, | ||
| ValueVector & | pos, | ||
| ValueVector & | ELcurr, | ||
| ValueVector & | ELideal, | ||
| RealType | start_phi0 | ||
| ) |
Minimize chi2 with respect to phi at zero for a fixed Rc.
| cusp | correction parameters |
| phiMO | uncorrected orbital (S-orbitals on this center only) |
| Z | nuclear charge |
| eta0 | value at zero for parts of the orbital that don't require correction - the non-S-orbitals on this center and all orbitals on other centers |
| pos | vector of radial positions |
| Elcurr | storage for current local energy |
| Elideal | storage for ideal local energy |
Definition at line 477 of file CuspCorrectionConstruction.cpp.
References APP_ABORT, bracket_minimum(), CuspCorrection::cparam, qmcplusplus::Units::charge::e, evaluateForPhi0Body(), find_minimum(), getIdealLocalEnergy(), getOriginalLocalEnergy(), getZeff(), ValGradLap::grad, ValGradLap::lap, OneMolecularOrbital::phi_vgl(), phiBar(), CuspCorrectionParameters::Rc, and ValGradLap::val.
Referenced by minimizeForRc().
| void minimizeForRc | ( | CuspCorrection & | cusp, |
| OneMolecularOrbital & | phiMO, | ||
| RealType | Z, | ||
| RealType | Rc_init, | ||
| RealType | Rc_max, | ||
| RealType | eta0, | ||
| ValueVector & | pos, | ||
| ValueVector & | ELcurr, | ||
| ValueVector & | ELideal | ||
| ) |
Minimize chi2 with respect to Rc and phi at zero.
| cusp | correction parameters |
| phiMO | uncorrected orbital (S-orbitals on this center only) |
| Z | nuclear charge |
| Rc_init | initial value for Rc |
| Rc_max | maximum value for Rc |
| eta0 | value at zero for parts of the orbital that don't require correction - the non-S-orbitals on this center and all orbitals on other centers |
| pos | vector of radial positions |
| Elcurr | storage for current local energy |
| Elideal | storage for ideal local energy |
Output is parameter values in cusp.cparam
Definition at line 524 of file CuspCorrectionConstruction.cpp.
References Bracket_min_t< T >::a, APP_ABORT, bracket_minimum(), CuspCorrection::cparam, qmcplusplus::Units::charge::e, find_minimum(), minimizeForPhiAtZero(), OneMolecularOrbital::phi(), CuspCorrectionParameters::Rc, and Bracket_min_t< T >::success.
Referenced by generateCuspInfo().
| void qmcplusplus::mpiTestFunctionWrapped | ( | Communicate * | comm, |
| std::vector< double > & | fake_args | ||
| ) |
Openmp generally works but is not guaranteed with std::atomic.
Definition at line 21 of file test_mpi_exception_wrapper.cpp.
References CHECK(), comm, and Communicate::size().
Referenced by TEST_CASE().
| void qmcplusplus::normalize | ( | ParticleAttrib< TinyVector< T, D >> & | pa | ) |
Definition at line 107 of file ParticleAttribOps.h.
| std::atomic<size_t> qmcplusplus::OMPallocator_device_mem_allocated | ( | 0 | ) |
| constexpr bool qmcplusplus::operator& | ( | DriverDebugChecks | x, |
| DriverDebugChecks | y | ||
| ) |
Definition at line 28 of file DriverDebugChecks.h.
|
inline |
Definition at line 121 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 131 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 203 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 218 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 269 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 285 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 302 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 357 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 378 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 422 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 451 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 519 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 556 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 589 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 628 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 661 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 693 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 726 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 769 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1007 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1208 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1616 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1854 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 2055 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 416 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 666 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 916 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1125 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1326 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1763 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1972 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 2173 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Definition at line 784 of file OhmmsMatrixOperators.h.
References evaluate().
|
inline |
Definition at line 817 of file ParticleAttrib.cpp.
References evaluate().
Definition at line 2281 of file OhmmsVectorOperators.h.
References evaluate().
|
inline |
Definition at line 192 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 554 of file TinyMatrixOps.h.
|
inline |
Definition at line 567 of file TinyMatrixOps.h.
|
inline |
Definition at line 576 of file TinyMatrixOps.h.
|
inline |
Definition at line 1539 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
| bool qmcplusplus::operator!= | ( | const CUDAManagedAllocator< T1 > & | , |
| const CUDAManagedAllocator< T2 > & | |||
| ) |
Definition at line 76 of file CUDAallocator.hpp.
| bool qmcplusplus::operator!= | ( | const SYCLSharedAllocator< T1 > & | , |
| const SYCLSharedAllocator< T2 > & | |||
| ) |
Definition at line 78 of file SYCLallocator.hpp.
| bool qmcplusplus::operator!= | ( | const Mallocator< T1, ALIGN1 > & | , |
| const Mallocator< T2, ALIGN2 > & | |||
| ) |
Definition at line 88 of file Mallocator.hpp.
| bool qmcplusplus::operator!= | ( | const CUDAAllocator< T1 > & | , |
| const CUDAAllocator< T2 > & | |||
| ) |
Definition at line 180 of file CUDAallocator.hpp.
|
inline |
Definition at line 180 of file CrystalLattice.cpp.
| bool qmcplusplus::operator!= | ( | const SYCLAllocator< T1 > & | , |
| const SYCLAllocator< T2 > & | |||
| ) |
Definition at line 181 of file SYCLallocator.hpp.
| bool qmcplusplus::operator!= | ( | const CUDAHostAllocator< T1 > & | , |
| const CUDAHostAllocator< T2 > & | |||
| ) |
Definition at line 230 of file CUDAallocator.hpp.
| bool qmcplusplus::operator!= | ( | const SYCLHostAllocator< T1 > & | , |
| const SYCLHostAllocator< T2 > & | |||
| ) |
Definition at line 245 of file SYCLallocator.hpp.
| bool qmcplusplus::operator!= | ( | const Matrix< T, Alloc > & | lhs, |
| const Matrix< T, Alloc > & | rhs | ||
| ) |
Definition at line 403 of file OhmmsMatrix.h.
|
inline |
Definition at line 405 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
| bool qmcplusplus::operator!= | ( | const Vector< T, Alloc > & | lhs, |
| const Vector< T, Alloc > & | rhs | ||
| ) |
Definition at line 438 of file OhmmsVector.h.
| bool qmcplusplus::operator!= | ( | const Tensor< T, D > & | lhs, |
| const Tensor< T, D > & | rhs | ||
| ) |
|
inline |
Definition at line 655 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 905 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1116 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1317 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1752 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1963 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 2164 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 110 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 118 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 192 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 206 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 258 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 274 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 290 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 347 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 368 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 412 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 441 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 508 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 545 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 618 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 683 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 758 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 997 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1198 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Definition at line 768 of file OhmmsMatrixOperators.h.
References evaluate().
|
inline |
Definition at line 801 of file ParticleAttrib.cpp.
References evaluate().
Definition at line 2265 of file OhmmsVectorOperators.h.
References evaluate().
|
inline |
Definition at line 98 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 105 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 180 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 194 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 235 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 262 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 278 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 338 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 358 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 403 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 431 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 485 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 533 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 609 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 674 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 735 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 979 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1180 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Definition at line 760 of file OhmmsMatrixOperators.h.
References evaluate().
|
inline |
Definition at line 793 of file ParticleAttrib.cpp.
References evaluate().
Definition at line 2249 of file OhmmsVectorOperators.h.
References evaluate().
|
inline |
Definition at line 38 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 45 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 74 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 79 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 157 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 168 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 170 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 212 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 239 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 254 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 320 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 337 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 385 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 410 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 462 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 474 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 495 of file AntiSymTensor.h.
|
inline |
Definition at line 510 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 591 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 656 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 712 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 961 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1162 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Definition at line 744 of file OhmmsMatrixOperators.h.
References evaluate().
|
inline |
Definition at line 777 of file ParticleAttrib.cpp.
References evaluate().
Definition at line 2233 of file OhmmsVectorOperators.h.
References evaluate().
|
inline |
Definition at line 31 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 37 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 85 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 92 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 161 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 168 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 182 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 223 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 250 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 266 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 329 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 347 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 394 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 420 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 467 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 473 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 521 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 600 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 665 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 723 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 970 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1171 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Definition at line 752 of file OhmmsMatrixOperators.h.
References evaluate().
|
inline |
Definition at line 785 of file ParticleAttrib.cpp.
References evaluate().
Definition at line 2241 of file OhmmsVectorOperators.h.
References evaluate().
concatenates two paths with a directory separator
| lhs | left-hand side of directory separator |
| rhs | right-hand side of directory separator |
Definition at line 75 of file hdf_path.cpp.
Definition at line 77 of file hdf_path.cpp.
Definition at line 79 of file hdf_path.cpp.
|
inline |
Definition at line 247 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 497 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 747 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 988 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1189 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1594 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1835 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 2036 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Definition at line 2257 of file OhmmsVectorOperators.h.
References evaluate().
|
inline |
Definition at line 350 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 600 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 850 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1071 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1272 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1697 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1918 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 2119 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
| std::ostream& qmcplusplus::operator<< | ( | std::ostream & | out, |
| const std::vector< T > & | rhs | ||
| ) |
collapsed vector printout [3, 3, 2, 2] is printed as [3(x2), 2(x2)]
Definition at line 25 of file StlPrettyPrint.hpp.
| std::ostream& qmcplusplus::operator<< | ( | std::ostream & | out, |
| const AxisGrid< T > & | rhs | ||
| ) |
Definition at line 29 of file test_ParseGridInput.cpp.
|
inline |
| std::ostream & operator<< | ( | std::ostream & | o_stream, |
| const VMCDriverInput & | vmci | ||
| ) |
Definition at line 34 of file VMCDriverInput.cpp.
|
inline |
Definition at line 46 of file NativeInitializerPrint.hpp.
| std::ostream & operator<< | ( | std::ostream & | out, |
| const DiracComputeBenchmarkParameters & | dcbmp | ||
| ) |
Definition at line 60 of file benchmark_DiracMatrixComputeCUDA.cpp.
References DiracComputeBenchmarkParameters::batch_size, DiracComputeBenchmarkParameters::n, and DiracComputeBenchmarkParameters::name.
| std::ostream & operator<< | ( | std::ostream & | out, |
| const testing::TestableNEReferencePoints & | rhs | ||
| ) |
Definition at line 50 of file test_ReferencePoints.cpp.
References TestableNEReferencePoints::write_testable_description().
|
inline |
Definition at line 57 of file NativeInitializerPrint.hpp.
|
inline |
Definition at line 68 of file NativeInitializerPrint.hpp.
| std::ostream & operator<< | ( | std::ostream & | o_stream, |
| const DMCDriverInput & | dmci | ||
| ) |
Definition at line 77 of file DMCDriverInput.cpp.
|
inline |
Definition at line 82 of file NativeInitializerPrint.hpp.
|
inline |
|
inline |
Definition at line 101 of file HDFVersion.h.
| std::ostream & operator<< | ( | std::ostream & | out, |
| const NEReferencePoints & | rhs | ||
| ) |
Definition at line 110 of file NEReferencePoints.cpp.
References NEReferencePoints::write_description().
|
inline |
Definition at line 127 of file ci_configuration2.h.
References ci_configuration2::occup.
|
inline |
Definition at line 128 of file ci_configuration.h.
References ci_configuration::occup.
| std::ostream& qmcplusplus::operator<< | ( | std::ostream & | o_stream, |
| const VMCBatched & | vmc_batched | ||
| ) |
|
inline |
Definition at line 185 of file string_utils.h.
References astring::s.
| std::ostream& qmcplusplus::operator<< | ( | std::ostream & | out, |
| const TinyVector< T, D > & | rhs | ||
| ) |
Definition at line 247 of file TinyVector.h.
References print().
| std::ostream & operator<< | ( | std::ostream & | os, |
| SFNBranch::VParamType & | rhs | ||
| ) |
Definition at line 322 of file SFNBranch.cpp.
| std::ostream& qmcplusplus::operator<< | ( | std::ostream & | out, |
| const OneDimGridBase< T > & | rhs | ||
| ) |
Definition at line 347 of file OneDimGridBase.h.
| std::ostream& qmcplusplus::operator<< | ( | std::ostream & | o_stream, |
| const QMCDriverNew & | qmcd | ||
| ) |
Definition at line 393 of file QMCDriverNew.cpp.
References app_log(), QMCDriverNew::current_step_, QMCDriverInput::get_max_blocks(), MCPopulation::get_num_local_walkers(), QMCDriverInput::get_sub_steps(), QMCDriverInput::get_tau(), QMCDriverNew::population_, QMCDriverNew::qmcdriver_input_, QMCDriverNew::steps_per_block_, and QMCDriverNew::target_samples_.
| std::ostream& qmcplusplus::operator<< | ( | std::ostream & | out, |
| const Matrix< T, Alloc > & | rhs | ||
| ) |
Definition at line 412 of file OhmmsMatrix.h.
|
inline |
Definition at line 438 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
| std::ostream& qmcplusplus::operator<< | ( | std::ostream & | out, |
| const Vector< T, Alloc > & | rhs | ||
| ) |
Definition at line 446 of file OhmmsVector.h.
| std::ostream& qmcplusplus::operator<< | ( | std::ostream & | out, |
| const SymTensor< T, D > & | rhs | ||
| ) |
Definition at line 449 of file SymTensor.h.
| std::ostream& qmcplusplus::operator<< | ( | std::ostream & | out, |
| const Tensor< T, D > & | rhs | ||
| ) |
| std::ostream& qmcplusplus::operator<< | ( | std::ostream & | out, |
| const AntiSymTensor< T, D > & | rhs | ||
| ) |
Definition at line 615 of file AntiSymTensor.h.
|
inline |
Definition at line 688 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
| std::ostream & operator<< | ( | std::ostream & | os, |
| SimpleFixedNodeBranch::VParamType & | rhs | ||
| ) |
Definition at line 822 of file SimpleFixedNodeBranch.cpp.
| std::ostream& qmcplusplus::operator<< | ( | std::ostream & | out, |
| const ParticleAttrib< T > & | rhs | ||
| ) |
Definition at line 922 of file ParticleAttrib.cpp.
|
inline |
Definition at line 938 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1143 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1785 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1990 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Definition at line 2297 of file OhmmsVectorOperators.h.
References evaluate().
|
inline |
Definition at line 361 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 611 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 861 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1080 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1281 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1708 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1927 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 2128 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
| bool qmcplusplus::operator== | ( | const CUDAManagedAllocator< T1 > & | , |
| const CUDAManagedAllocator< T2 > & | |||
| ) |
Definition at line 71 of file CUDAallocator.hpp.
| bool qmcplusplus::operator== | ( | const SYCLSharedAllocator< T1 > & | , |
| const SYCLSharedAllocator< T2 > & | |||
| ) |
Definition at line 73 of file SYCLallocator.hpp.
Checks whether lhs and rhs are equal.
| lhs | first path |
| rhs | second path |
Definition at line 81 of file hdf_path.cpp.
| bool qmcplusplus::operator== | ( | const Mallocator< T1, ALIGN1 > & | , |
| const Mallocator< T2, ALIGN2 > & | |||
| ) |
Definition at line 83 of file Mallocator.hpp.
|
inline |
Definition at line 171 of file CrystalLattice.cpp.
References abs(), and CrystalLattice< T, D >::R.
| bool qmcplusplus::operator== | ( | const CUDAAllocator< T1 > & | , |
| const CUDAAllocator< T2 > & | |||
| ) |
Definition at line 175 of file CUDAallocator.hpp.
| bool qmcplusplus::operator== | ( | const SYCLAllocator< T1 > & | , |
| const SYCLAllocator< T2 > & | |||
| ) |
Definition at line 176 of file SYCLallocator.hpp.
Definition at line 191 of file string_utils.h.
References astring::s.
| bool qmcplusplus::operator== | ( | const CUDAHostAllocator< T1 > & | , |
| const CUDAHostAllocator< T2 > & | |||
| ) |
Definition at line 225 of file CUDAallocator.hpp.
| bool qmcplusplus::operator== | ( | const SYCLHostAllocator< T1 > & | , |
| const SYCLHostAllocator< T2 > & | |||
| ) |
Definition at line 239 of file SYCLallocator.hpp.
Definition at line 388 of file OhmmsMatrix.h.
References Matrix< T, Alloc >::size().
|
inline |
Definition at line 394 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Definition at line 423 of file OhmmsVector.h.
References Vector< T, Alloc >::size().
|
inline |
Definition at line 644 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 894 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1107 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1308 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1741 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1954 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 2155 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 372 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 622 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 872 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1089 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1290 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1719 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1936 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 2137 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 383 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 633 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 883 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1098 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1299 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1730 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1945 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 2146 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Read unit type recorded in int.
This should really be human readable TODO: support both until going to string only.
Definition at line 49 of file PosUnit.h.
|
inline |
Definition at line 107 of file HDFVersion.h.
|
inline |
Definition at line 177 of file string_utils.h.
References astring::s.
| std::istream& qmcplusplus::operator>> | ( | std::istream & | is, |
| TinyVector< T, D > & | rhs | ||
| ) |
Definition at line 254 of file TinyVector.h.
| std::istream& qmcplusplus::operator>> | ( | std::istream & | is, |
| Matrix< T, Alloc > & | rhs | ||
| ) |
Definition at line 427 of file OhmmsMatrix.h.
References Matrix< T, Alloc >::size().
|
inline |
Definition at line 450 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
| std::istream& qmcplusplus::operator>> | ( | std::istream & | is, |
| Vector< T, Alloc > & | rhs | ||
| ) |
Definition at line 455 of file OhmmsVector.h.
References Vector< T, Alloc >::size().
| std::istream& qmcplusplus::operator>> | ( | std::istream & | is, |
| Tensor< T, D > & | rhs | ||
| ) |
|
inline |
Definition at line 700 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 950 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1153 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1797 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 2000 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Definition at line 2305 of file OhmmsVectorOperators.h.
References evaluate().
|
inline |
Definition at line 145 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 157 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 227 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 242 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 293 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 309 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 326 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 376 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 400 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 441 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 473 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 543 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 580 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 647 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 712 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 793 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1026 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1227 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Definition at line 792 of file OhmmsMatrixOperators.h.
References evaluate().
|
inline |
Definition at line 825 of file ParticleAttrib.cpp.
References evaluate().
Definition at line 2289 of file OhmmsVectorOperators.h.
References evaluate().
| constexpr DriverDebugChecks qmcplusplus::operator| | ( | DriverDebugChecks | x, |
| DriverDebugChecks | y | ||
| ) |
Definition at line 33 of file DriverDebugChecks.h.
|
inline |
Definition at line 133 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 144 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 215 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 230 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 281 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 297 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 314 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 366 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 389 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 431 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 462 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 531 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 568 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 637 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 702 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 781 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1016 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1217 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 40 of file DriverDebugChecks.h.
Definition at line 776 of file OhmmsMatrixOperators.h.
References evaluate().
|
inline |
Definition at line 809 of file ParticleAttrib.cpp.
References evaluate().
Definition at line 2273 of file OhmmsVectorOperators.h.
References evaluate().
|
inline |
Definition at line 427 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 677 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 927 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1134 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1335 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1774 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1981 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 2182 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
| constexpr DriverDebugChecks qmcplusplus::operator~ | ( | DriverDebugChecks | x | ) |
Definition at line 38 of file DriverDebugChecks.h.
|
inline |
Definition at line 47 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 53 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 177 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 483 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
| OneBodyDensityMatrices qmcplusplus::original | ( | std::move(obdmi) | , |
| pset_target. | getLattice(), | ||
| species_set | , | ||
| spomap | , | ||
| pset_target | |||
| ) |
|
inline |
Definition at line 211 of file TinyVector.h.
Referenced by DiracDeterminantWithBackflow::dummyEvalLi(), Backflow_eI< FT >::evaluate(), Backflow_ee< FT >::evaluate(), DiracDeterminantWithBackflow::evaluateDerivatives(), J1Spin< FT >::evaluateHessian(), DiracDeterminant< DU_TYPE >::evaluateHessian(), TwoBodyJastrow< FT >::evaluateHessian(), J1OrbitalSoA< FT >::evaluateHessian(), DiracDeterminantBatched< PL, VT, FPVT >::evaluateHessian(), StressPBC::evaluateKineticSymTensor(), DiracDeterminantWithBackflow::evaluateLog(), Backflow_eI< FT >::evaluatePbyP(), Backflow_ee< FT >::evaluatePbyP(), Backflow_eI< FT >::evaluateWithDerivatives(), and Backflow_ee< FT >::evaluateWithDerivatives().
| void qmcplusplus::output_vector | ( | const std::string & | name, |
| std::vector< int > & | vec | ||
| ) |
Definition at line 33 of file test_walker_control.cpp.
| void qmcplusplus::pad_string | ( | const std::string & | in, |
| std::string & | out, | ||
| int | field_len | ||
| ) |
Definition at line 351 of file TimerManager.cpp.
Referenced by TimerManager< qmcplusplus::TimerType< CLOCK > >::print_stack().
| void qmcplusplus::parse_electron_ion_pbc_z | ( | ParticleSet & | ions, |
| ParticleSet & | electrons | ||
| ) |
Definition at line 382 of file test_distance_table.cpp.
References doc, OhmmsElementBase::getName(), Libxml2Document::getRoot(), ParticleSet::isSameMass(), okay, Libxml2Document::parseFromString(), XMLParticleParser::readXML(), and REQUIRE().
Referenced by TEST_CASE(), test_distance_fcc_pbc_z_batched_APIs(), test_distance_pbc_z_batched_APIs(), and test_distance_pbc_z_batched_APIs_ee_NEED_TEMP_DATA_ON_HOST().
| SimulationCell qmcplusplus::parse_pbc_fcc_lattice | ( | ) |
Definition at line 316 of file test_distance_table.cpp.
References app_log(), doc, Libxml2Document::getRoot(), lattice, okay, Libxml2Document::parseFromString(), LatticeParser::put(), and REQUIRE().
Referenced by test_distance_fcc_pbc_z_batched_APIs().
| SimulationCell qmcplusplus::parse_pbc_lattice | ( | ) |
Definition at line 349 of file test_distance_table.cpp.
References app_log(), doc, Libxml2Document::getRoot(), lattice, okay, Libxml2Document::parseFromString(), LatticeParser::put(), and REQUIRE().
Referenced by TEST_CASE(), test_distance_pbc_z_batched_APIs(), and test_distance_pbc_z_batched_APIs_ee_NEED_TEMP_DATA_ON_HOST().
| AxisGrid< REAL > parseGridInput | ( | std::istringstream & | grid_input_stream | ) |
Parses the one dimensional grid specification format originated for EnergyDensity This has be refactored from QMCHamiltonian/SpaceGrid.cpp.
My evidence for correctness is that it can pass the unit test I wrote, there was no legacy test.
Definition at line 23 of file ParseGridInput.cpp.
References abs(), AxisGrid< REAL >::dimensions, AxisGrid< REAL >::du_int, qmcplusplus::Units::charge::e, floor(), AxisGrid< REAL >::gmap, omptarget::min(), n, AxisGrid< REAL >::ndom_int, AxisGrid< REAL >::ndu_int, AxisGrid< REAL >::ndu_per_interval, AxisGrid< REAL >::odu, qmcplusplus::modernstrutil::split(), AxisGrid< REAL >::umax, and AxisGrid< REAL >::umin.
| template AxisGrid< double > parseGridInput< double > | ( | std::istringstream & | grid_input_stream | ) |
| template AxisGrid< float > parseGridInput< float > | ( | std::istringstream & | grid_input_stream | ) |
| template void qmcplusplus::ParticleSet::mw_accept_rejectMove< CoordsType::POS > | ( | const RefVectorWithLeader< ParticleSet > & | p_list, |
| Index_t | iat, | ||
| const std::vector< bool > & | isAccepted, | ||
| bool | forward_mode | ||
| ) |
| template void qmcplusplus::ParticleSet::mw_accept_rejectMove< CoordsType::POS_SPIN > | ( | const RefVectorWithLeader< ParticleSet > & | p_list, |
| Index_t | iat, | ||
| const std::vector< bool > & | isAccepted, | ||
| bool | forward_mode | ||
| ) |
| template void qmcplusplus::ParticleSet::mw_makeMove< CoordsType::POS > | ( | const RefVectorWithLeader< ParticleSet > & | p_list, |
| Index_t | iat, | ||
| const MCCoords< CoordsType::POS > & | displs | ||
| ) |
| template void qmcplusplus::ParticleSet::mw_makeMove< CoordsType::POS_SPIN > | ( | const RefVectorWithLeader< ParticleSet > & | p_list, |
| Index_t | iat, | ||
| const MCCoords< CoordsType::POS_SPIN > & | displs | ||
| ) |
|
inline |
Definition at line 55 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 61 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 185 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 491 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 62 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 69 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 200 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 498 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 127 of file Combiners.h.
References Combine1< A, Op, Tag >::combine().
|
inline |
Definition at line 133 of file Combiners.h.
|
inline |
Definition at line 139 of file Combiners.h.
References Combine3< A, B, C, Op, Tag >::combine().
| RealType phiBar | ( | const CuspCorrection & | cusp, |
| RealType | r, | ||
| OneMolecularOrbital & | phiMO | ||
| ) |
Definition at line 364 of file CuspCorrectionConstruction.cpp.
References CuspCorrectionParameters::C, CuspCorrection::cparam, OneMolecularOrbital::phi(), CuspCorrectionParameters::Rc, and CuspCorrection::Rr().
Referenced by computeRadialPhiBar(), evaluateForPhi0Body(), getCurrentLocalEnergy(), and minimizeForPhiAtZero().
| std::vector<int, CUDAHostAllocator<int> > qmcplusplus::pivots | ( | 8 | , |
| -1. | 0 | ||
| ) |
| void qmcplusplus::PosAoS2SoA | ( | int | nrows, |
| int | ncols, | ||
| const T *restrict | iptr, | ||
| int | lda, | ||
| T *restrict | out, | ||
| int | ldb | ||
| ) |
General conversion function from AoS[nrows][ncols] to SoA[ncols][ldb].
| nrows | the first dimension |
| ncols | the second dimension |
| iptr | input pointer |
| lda | stride of iptr |
| out | output pointer |
| lda | strided of out |
Modeled after blas/lapack for lda/ldb
Definition at line 106 of file PosTransformer.h.
Referenced by VectorSoaContainer< ST, 5 >::copyIn().
| void qmcplusplus::PosSoA2AoS | ( | int | nrows, |
| int | ncols, | ||
| const T *restrict | iptr, | ||
| int | lda, | ||
| T *restrict | out, | ||
| int | ldb | ||
| ) |
General conversion function from SoA[ncols][ldb] to AoS[nrows][ncols].
| nrows | the first dimension |
| ncols | the second dimension |
| iptr | input pointer |
| lda | stride of iptr |
| out | output pointer |
| lda | strided of out |
Modeled after blas/lapack for lda/ldb
Definition at line 133 of file PosTransformer.h.
References lda.
Referenced by VectorSoaContainer< ST, 5 >::copyOut().
|
inline |
|
inline |
Definition at line 316 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by GaussianCombo< T >::addGaussian(), BackflowBuilder::addRPA(), LRRPAHandlerTemp< Func, BreakupBasis >::Breakup(), RPAJastrow::buildOrbital(), LPQHISRCoulombBasis::c(), CartesianTensor< T, Point_t, Tensor_t, GGG_t >::CartesianTensor(), SimpleFixedNodeBranch::collect(), ACForce::compute_regularizer_f(), QMCCostFunction::correlatedSampling(), QMCCostFunctionBatched::correlatedSampling(), RadialJastrowBuilder::createJ1(), LPQHISRCoulombBasis::dc_dk(), SpaceWarpTransformation::df(), GaussianTimesRN< T >::BasicGaussian::df(), GenericSTO< T >::df(), RadialSTO< T >::df(), LPQHISRCoulombBasis::dh_dr(), LPQHISRCoulombBasis::Eminus_dG(), ForceCeperley::evaluate(), GaussianTimesRN< T >::BasicGaussian::evaluate(), UserFunctor< T >::evaluate(), GenericSTO< T >::evaluate(), RadialSTO< T >::evaluate(), ExampleHeComponent::evaluateDerivatives(), UserFunctor< T >::evaluateDerivatives(), qmcplusplus::ewaldref::ewaldSum(), SpaceWarpTransformation::f(), GaussianTimesRN< T >::BasicGaussian::f(), GenericSTO< T >::f(), RadialSTO< T >::f(), LRRPABFeeHandlerTemp< Func, BreakupBasis >::fillFk(), LRRPAHandlerTemp< Func, BreakupBasis >::fillFk(), ForceChiesaPBCAA::g_filter(), GaussianCombo< T >::GaussianCombo(), HEGGrid< T >::getCellLength(), qmcplusplus::ewaldref::getKappaEwald(), qmcplusplus::ewaldref::getKappaMadelung(), HEGGrid< T >::getNC(), QMCCostFunction::GradCost(), QMCCostFunctionBatched::GradCost(), LPQHISRCoulombBasis::h(), LPQHISRCoulombBasis::hintr2(), LogGrid::Init(), LRRPABFeeHandlerTemp< Func, BreakupBasis >::InitBreakup(), LRRPAHandlerTemp< Func, BreakupBasis >::InitBreakup(), LRHandlerTemp< Func, BreakupBasis >::InitBreakup(), LRHandlerSRCoulomb< Func, BreakupBasis >::InitBreakup(), QMCFiniteSize::initialize(), ForceCeperley::InitMatrix(), ForceChiesaPBCAA::InitMatrix(), ForceBase::initVarReduction(), LogGrid::LogGrid(), qmcplusplus::ewaldref::madelungSum(), MomentumDistribution::MomentumDistribution(), DensityMatrices1B::normalize(), OneBodyDensityMatrices::normalizeBasis(), OneBodyDensityMatrices::OneBodyDensityMatrices(), STONorm< T >::operator()(), FnPow::operator()(), GaussianCombo< T >::putBasisGroupH5(), MomentumEstimator::putSpecial(), YukawaBreakup< T >::reset(), DerivRPABreakup< T >::reset(), RPABreakup< T >::reset(), DerivYukawaBreakup< T >::reset(), EPRPABreakup< T >::reset(), derivEPRPABreakup< T >::reset(), RPABFeeBreakup< T >::reset(), LPQHISRCoulombBasis::rh(), PairCorrEstimator::set_norm_factor(), DensityMatrices1B::set_state(), SoaCartesianTensor< T >::SoaCartesianTensor(), SoaSphericalTensor< ST >::SoaSphericalTensor(), SphericalTensor< T, Point_t, Tensor_t, GGG_t >::SphericalTensor(), TEST_CASE(), DensityMatrices1B::test_overlap(), OneBodyDensityMatricesTests< T >::testGenerateSamples(), SHOSetBuilder::update_basis_states(), DescentEngine::updateParameters(), DerivRPABreakup< T >::Xk(), RPABreakup< T >::Xk(), EPRPABreakup< T >::Xk(), derivEPRPABreakup< T >::Xk(), RPABFeeBreakup< T >::Xk(), and Ylm().
|
inline |
Definition at line 566 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 816 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1044 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1245 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1663 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1891 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 2092 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
| void print | ( | OptimizableFunctorBase & | func, |
| std::ostream & | os, | ||
| double | extent = -1.0 |
||
| ) |
evaluates a functor (value and derivative) and dumps the quantities to output
| func | the functor for which the value and derivative is evaluated |
| os | the output stream to write the quantities to |
| extent | the functor is evaluated from [0, extent) unless extent < 0, in which case the functor is evaluated from [0, cutoff_radius) |
Definition at line 17 of file OptimizableFunctorBase.cpp.
References OptimizableFunctorBase::cutoff_radius, OptimizableFunctorBase::df(), OptimizableFunctorBase::f(), and n.
Referenced by RadialJastrowBuilder::createJ1(), RadialJastrowBuilder::createJ2(), and operator<<().
| void print_mem | ( | const std::string & | title, |
| std::ostream & | log | ||
| ) |
Definition at line 30 of file MemoryUsage.cpp.
References freemem(), getCUDAdeviceFreeMem(), getCUDAdeviceMemAllocated(), getOMPdeviceMemAllocated(), getSYCLdeviceFreeMem(), getSYCLdeviceMemAllocated(), log(), and memusage().
Referenced by CSUpdateBase::initCSWalkersForPbyP(), QMCUpdateBase::initWalkersForPbyP(), main(), CloneManager::makeClones(), VMCBatched::process(), DMCBatched::process(), QMCMain::QMCMain(), VMCBatched::run(), DMCBatched::run(), and TEST_CASE().
| void qmcplusplus::print_vector | ( | vector< int > & | out | ) |
Definition at line 25 of file test_partition.cpp.
|
inline |
Find a better place for other user classes, Matrix should be padded as well.
Definition at line 193 of file LCAOrbitalSet.cpp.
References qmcplusplus::Units::distance::A, B(), qmcplusplus::Units::charge::C, BLAS::gemm(), and BLAS::zone.
Referenced by LCAOrbitalSet::evaluate_notranspose(), LCAOrbitalSet::evaluateGradSource(), LCAOrbitalSet::evaluateGradSourceRow(), LCAOrbitalSet::evaluateVGH(), LCAOrbitalSet::evaluateVGHGH(), and LCAOrbitalSet::evaluateVGL().
| template void qmcplusplus::PSdispatcher::flex_accept_rejectMove< CoordsType::POS > | ( | const RefVectorWithLeader< ParticleSet > & | p_list, |
| int | iat, | ||
| const std::vector< bool > & | isAccepted, | ||
| bool | forward_mode | ||
| ) | const |
| template void qmcplusplus::PSdispatcher::flex_accept_rejectMove< CoordsType::POS_SPIN > | ( | const RefVectorWithLeader< ParticleSet > & | p_list, |
| int | iat, | ||
| const std::vector< bool > & | isAccepted, | ||
| bool | forward_mode | ||
| ) | const |
| template void qmcplusplus::PSdispatcher::flex_makeMove< CoordsType::POS > | ( | const RefVectorWithLeader< ParticleSet > & | p_list, |
| int | iat, | ||
| const MCCoords< CoordsType::POS > & | displs | ||
| ) | const |
| template void qmcplusplus::PSdispatcher::flex_makeMove< CoordsType::POS_SPIN > | ( | const RefVectorWithLeader< ParticleSet > & | p_list, |
| int | iat, | ||
| const MCCoords< CoordsType::POS_SPIN > & | displs | ||
| ) | const |
|
inline |
inout[i]=inout[i]-floor(inout[i])
See CPU/SIMD/vmath.h and should be specialized for vector libraries, e.g., INTEL vml, IBM massv
Definition at line 332 of file FastParticleOperators.h.
References qmcplusplus::simd::remainder().
Referenced by ParticleSet::convert2UnitInBox().
|
inline |
out[i]=in[i]-floor(in[i])
Definition at line 340 of file FastParticleOperators.h.
References qmcplusplus::simd::remainder().
|
inline |
Definition at line 23 of file kSpaceJastrowBuilder.cpp.
Referenced by kSpaceJastrowBuilder::buildComponent().
| std::unique_ptr< QMCFixedSampleLinearOptimizeBatched > QMCWFOptLinearFactoryNew | ( | xmlNodePtr | cur, |
| const ProjectData & | project_data, | ||
| const std::optional< EstimatorManagerInput > & | global_emi, | ||
| WalkerConfigurations & | wc, | ||
| MCPopulation && | pop, | ||
| SampleStack & | samples, | ||
| Communicate * | comm | ||
| ) |
Definition at line 10 of file QMCWFOptFactoryNew.cpp.
References app_summary(), comm, qmcplusplus::Units::charge::e, VMCDriverInput::readXML(), and QMCDriverInput::readXML().
Referenced by QMCDriverFactory::createQMCDriver().
|
inline |
Definition at line 1369 of file WaveFunctionTester.cpp.
References assignUniformRand(), and Random.
Referenced by WaveFunctionTester::runRatioV().
| void qmcplusplus::randomUpdateAccumulate | ( | testing::RandomForTest< QMCT::RealType > & | rft, |
| UPtrVector< OperatorEstBase > & | crowd_sdns | ||
| ) |
Definition at line 89 of file test_SpinDensityNew.cpp.
References SpinDensityNew::accumulate(), QMCTraits::DIM, RandomForTest< VT >::fillVecRng(), pset, and qmcplusplus::hdf::walkers.
| bool readCuspInfo | ( | const std::string & | cuspInfoFile, |
| const std::string & | objectName, | ||
| int | OrbitalSetSize, | ||
| Matrix< CuspCorrectionParameters > & | info | ||
| ) |
Read cusp correction parameters from XML file.
Definition at line 30 of file CuspCorrectionConstruction.cpp.
References OhmmsAttributeSet::add(), APP_ABORT, app_log(), getNodeName(), Libxml2Document::getRoot(), Libxml2Document::parse(), OhmmsAttributeSet::put(), and Matrix< T, Alloc >::rows().
Referenced by LCAOrbitalBuilder::createSPOSetFromXML(), and TEST_CASE().
| void qmcplusplus::readNameTypeLikeLegacy | ( | InputSection & | input_section_, |
| std::string & | name, | ||
| std::string & | type | ||
| ) |
make input class state consistent with legacy ignoring the distinction between the name and type of estimator for scalar estimators.
| [in] | input_section_ | naming here is to allow reuse of LAMBDA_setIfInInput macro |
| [in,out] | name | output param but assumed to be unset |
| [in,out] | type | output param but assumed to be unset |
Definition at line 26 of file ScalarEstimatorInputs.cpp.
References LAMBDA_setIfInInput.
Referenced by CSLocalEnergyInput::CSLocalEnergyInput(), LocalEnergyInput::LocalEnergyInput(), and RMCLocalEnergyInput::RMCLocalEnergyInput().
|
inline |
real part of a scalar. Cannot be replaced by std::real due to AFQMC specific needs.
Definition at line 86 of file complex_help.hpp.
Referenced by accum_sample(), SelfHealingOverlap::accumulate(), OTCDot< T1, T2, D >::apply(), OTCDot_CC< T1, T2, D >::apply(), OTAssign< TinyVector< T1, 3 >, TinyVector< std::complex< T2 >, 3 >, OP >::apply(), Quadrature3D< T >::CheckQuadratureRule(), DensityMatrices1B::compare(), MPC::compute_g_G(), compute_norm(), Copy(), kSpaceJastrow::evalGrad(), SelfHealingOverlapLegacy::evaluate(), PWRealOrbitalSet::evaluate(), DensityMatrices1B::evaluate_loop(), DensityMatrices1B::evaluate_matrix(), PWRealOrbitalSet::evaluate_notranspose(), BareKineticEnergy::evaluate_sp(), DiracDeterminantWithBackflow::evaluateDerivatives(), kSpaceJastrow::evaluateDerivatives(), kSpaceJastrow::evaluateLog(), OneBodyDensityMatrices::evaluateMatrix(), kSpaceJastrow::evaluateRatiosAlltoOne(), PWRealOrbitalSet::evaluateValue(), PWRealOrbitalSet::evaluateVGL(), ComplexExpFitClass< M >::Fit(), ComplexExpFitClass< M >::FitCusp(), fix_phase_rotate(), MPC::init_spline(), MomentumDistribution::MomentumDistribution(), BareKineticEnergy::mw_evaluatePerParticle(), DensityMatrices1B::normalize(), OneBodyDensityMatrices::normalizeBasis(), kSpaceJastrow::ratio(), kSpaceJastrow::ratioGrad(), WaveFunctionTester::runZeroVarianceTest(), TEST_CASE(), SHOSet::test_derivatives(), DiracDeterminantWithBackflow::testDerivFjj(), DiracDeterminantWithBackflow::testGGG(), and OrbitalImages::write_orbital_xsf().
|
inline |
Definition at line 87 of file complex_help.hpp.
|
inline |
Definition at line 88 of file complex_help.hpp.
|
inline |
Definition at line 89 of file complex_help.hpp.
|
inline |
Definition at line 126 of file string_utils.h.
| void removeSTypeOrbitals | ( | const std::vector< bool > & | corrCenter, |
| LCAOrbitalSet & | Phi | ||
| ) |
Remove S atomic orbitals from all molecular orbitals on all centers.
Definition at line 256 of file CuspCorrectionConstruction.cpp.
References LCAOrbitalSet::C, LCAOrbitalSet::getBasisSetSize(), SPOSet::getOrbitalSetSize(), and LCAOrbitalSet::myBasisSet.
Referenced by applyCuspCorrection(), and TEST_CASE().
| qmcplusplus::REQUIRE | ( | std::filesystem::exists(filename) | ) |
Referenced by check_force_copy(), check_matrix(), complex_test_case(), create_CN_Hamiltonian(), create_CN_particlesets(), qmcplusplus::testing::createEstimatorManagerNewGlobalInputXML(), qmcplusplus::testing::createEstimatorManagerNewInputXML(), qmcplusplus::testing::createEstimatorManagerNewVMCInputXML(), doSOECPotentialTest(), double_test_case(), MinTest::find_bracket(), MinTest::find_bracket_bound(), MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalWaveFunctionPool::make_O2_spinor(), MinimalWaveFunctionPool::make_O2_spinor_J12(), parse_electron_ion_pbc_z(), parse_pbc_fcc_lattice(), parse_pbc_lattice(), setup_He_wavefunction(), setupParticleSetPool(), setupParticleSetPoolBe(), test_C_diamond(), test_cartesian_ao(), optimize::TEST_CASE(), TEST_CASE(), TEST_CASE(), test_diamond_2x1x1_xml_input(), test_dirac_ao(), test_EtOH_mw(), test_HCN(), test_hcpBe_rotation(), test_He(), test_He_mw(), test_He_sto3g_xml_input(), test_J1_spline(), test_J3_polynomial3D(), test_LCAO_DiamondC_2x1x1_cplx(), test_LCAO_DiamondC_2x1x1_real(), test_lcao_spinor(), test_lcao_spinor_excited(), test_lcao_spinor_ion_derivs(), test_LiH_msd(), test_LiH_msd_xml_input(), test_LiH_msd_xml_input_with_positron(), test_Ne(), test_real_accumulator_basic(), test_spline_bounds(), test_tiny_vector(), OneBodyDensityMatricesTests< T >::testRegisterAndWrite(), and testTrialWaveFunction_diamondC_2x1x1().
| qmcplusplus::REQUIRE | ( | std::filesystem::remove(filename) | ) |
| REQUIRE | ( | okay | ) |
| REQUIRE | ( | clone ! | = nullptr | ) |
| REQUIRE | ( | clone.get() ! | = &original | ) |
| REQUIRE | ( | dynamic_cast< decltype(&original)>(clone.get()) ! | = nullptr | ) |
| void qmcplusplus::reset_string_output | ( | ) |
Definition at line 25 of file test_output_manager.cpp.
References app_out, debug_out, err_out, and summary_out.
Referenced by init_string_output(), and TEST_CASE().
| void saveCusp | ( | const std::string & | filename, |
| const Matrix< CuspCorrectionParameters > & | info, | ||
| const std::string & | id | ||
| ) |
save cusp correction info to a file.
Definition at line 130 of file CuspCorrectionConstruction.cpp.
References app_log(), qmcplusplus::Units::charge::C, Matrix< T, Alloc >::cols(), doc, and Matrix< T, Alloc >::rows().
Referenced by LCAOrbitalBuilder::createSPOSetFromXML().
| SpinDensityInput qmcplusplus::sdi | ( | node | ) |
| void qmcplusplus::set_num_calls | ( | TimerType< CLOCK > * | timer, |
| long | num_calls_input | ||
| ) |
Definition at line 32 of file test_timer.cpp.
References TimerType< CLOCK >::num_calls.
Referenced by TEST_CASE().
| void qmcplusplus::set_total_time | ( | TimerType< CLOCK > * | timer, |
| double | total_time_input | ||
| ) |
Definition at line 26 of file test_timer.cpp.
References TimerType< CLOCK >::total_time.
Referenced by TEST_CASE().
|
inline |
da = scaled(tau)*ga
| tau | time step |
| qf | real quantum forces |
| drift | drift |
Definition at line 166 of file DriftOperators.h.
References getDriftScale(), and qmcplusplus::Units::time::s.
Referenced by RMCUpdateAllWithDrift::advanceWalkersRMC(), and RMCUpdateAllWithDrift::advanceWalkersVMC().
|
inline |
da = scaled(tau)*ga
| tau | time step |
| qf | real quantum forces |
| drift | drift |
Definition at line 178 of file DriftOperators.h.
References getDriftScale(), and qmcplusplus::Units::time::s.
|
inline |
da = scaled(tau)*ga
| tau | time step |
| qf | complex quantum forces |
| drift | drift |
Definition at line 191 of file DriftOperators.h.
References convertToReal(), getDriftScale(), and qmcplusplus::Units::time::s.
|
inline |
scale drift
| tau_au | timestep au |
| qf | quantum forces |
| drift | scaled quantum forces |
| return | correction term |
Assume, mass=1
Definition at line 86 of file DriftOperators.h.
References convertToReal(), dot(), norm(), and sqrt().
Referenced by DMCUpdateAllWithRejection::advanceWalker(), DMCUpdateAllWithKill::advanceWalker(), RMCUpdateAllWithDrift::advanceWalkersVMC(), QMCUpdateBase::getNodeCorrection(), RMCUpdateAllWithDrift::initWalkers(), QMCUpdateBase::initWalkers(), and TEST_CASE().
|
inline |
scale drift
| tau_au | timestep au |
| massinv | 1/m per particle |
| qf | quantum forces, i.e. grad_psi_over_psi |
| drift | scaled importance-sampling drift |
| return | correction term |
Fill the drift vector one particle at a time (pbyp).
The naive drift is tau/mass*|grad_psi_over_psi|, grad_psi_over_psi the log derivative of the guiding wavefunction; tau is timestep; mass is particle mass The naive drift diverges at a node and is large near a nucleus. Both cases may cause persistent configurations, because the reverse proposal probability is virtually zero.
The norm of the drift vector should be limited in two ways:
T should be either float or double T1 may be real (float or double) or complex<float or double> D should be the number of spatial dimensions (int)
Definition at line 131 of file DriftOperators.h.
References convertToReal(), dot(), norm(), and sqrt().
| void qmcplusplus::setSpeciesProperty | ( | SpeciesSet & | tspecies, |
| int | sid, | ||
| xmlNodePtr | cur | ||
| ) |
set the property of a SpeciesSet
| tspecies | SpeciesSet |
| sid | id of the species whose properties to be set |
Need unit handlers but for now, everything is in AU: m_e=1.0, hartree and bohr and unit="au" Example to define C(arbon) <group name="C"> <parameter name="mass" unit="amu">12</parameter> <parameter name="charge">-6</parameter> </group> Note that unit="amu" is given for the mass. When mass is not given, they are set to the electron mass.
Definition at line 46 of file XMLParticleIO.cpp.
References OhmmsAttributeSet::add(), SpeciesSet::addAttribute(), qmcplusplus::Units::proton_mass, OhmmsAttributeSet::put(), and putContent().
Referenced by XMLParticleParser::readXML(), and XMLParticleParser::reset().
| std::unique_ptr<TrialWaveFunction> qmcplusplus::setup_He_wavefunction | ( | Communicate * | c, |
| ParticleSet & | elec, | ||
| ParticleSet & | ions, | ||
| const WaveFunctionFactory::PSetMap & | particle_set_map | ||
| ) |
Definition at line 30 of file test_TrialWaveFunction_He.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), WaveFunctionFactory::buildTWF(), ParticleSet::create(), doc, Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), and ParticleSet::resetGroups().
Referenced by TEST_CASE().
| SetupSFNBranch qmcplusplus::setup_sfnb | ( | pools. | comm | ) |
| void setupParticleSetPool | ( | ParticleSetPool & | pp | ) |
Definition at line 36 of file test_RotatedSPOs_LCAO.cpp.
References doc, Libxml2Document::getRoot(), okay, Libxml2Document::parseFromString(), ParticleSetPool::put(), and REQUIRE().
Referenced by TEST_CASE().
| void qmcplusplus::setupParticleSetPoolBe | ( | ParticleSetPool & | pp | ) |
Definition at line 78 of file test_RotatedSPOs_LCAO.cpp.
References doc, Libxml2Document::getRoot(), okay, Libxml2Document::parseFromString(), ParticleSetPool::put(), and REQUIRE().
Referenced by TEST_CASE().
| std::string qmcplusplus::setupRotationXML | ( | const std::string & | rot_angle_up, |
| const std::string & | rot_angle_down, | ||
| const std::string & | coeff_up, | ||
| const std::string & | coeff_down | ||
| ) |
Definition at line 127 of file test_RotatedSPOs_LCAO.cpp.
Referenced by TEST_CASE().
|
inline |
Definition at line 120 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by assignGaussRand(), QMCFiniteSize::build_spherical_grid(), LatticeAnalyzer< T, 2 >::calcSimulationCellRadius(), RadialJastrowBuilder::computeJ2uk(), J2KECorrection< RealType, FT >::computeKEcorr(), LPQHI_BasisClass::dEminus_dk(), LPQHI_BasisClass::dEplus_dk(), derivYlmSpherical(), LPQHIBasis::Eminus(), LPQHISRCoulombBasis::Eminus(), LPQHI_BasisClass::Eminus(), LPQHISRCoulombBasis::Eminus_dG(), LPQHIBasis::Eplus(), LPQHISRCoulombBasis::Eplus(), LPQHI_BasisClass::Eplus(), LPQHISRCoulombBasis::Eplus_dG(), SpinorSet::evaluate_notranspose(), SpinorSet::evaluate_notranspose_spin(), SpinorSet::evaluate_spin(), SpinorSet::evaluateDetSpinorRatios(), SpinorSet::evaluateGradSource(), SpinorSet::evaluateValue(), SpinorSet::evaluateVGL(), SpinorSet::evaluateVGL_spin(), generateRotationMatrix(), MagnetizationDensity::generateSpinIntegrand(), MPC::init_f_G(), SpaceGrid::initialize_rectilinear(), ChannelPotential::jl(), kSpaceJastrow::kSpaceJastrow(), SOECPComponent::matrixElementDecomposed(), SpinorSet::mw_evaluate_notranspose(), SpinorSet::mw_evaluateVGLandDetRatioGradsWithSpin(), SpinorSet::mw_evaluateVGLWithSpin(), FnSin::operator()(), Quadrature3D< T >::Quadrature3D(), WaveFunctionTester::runBasicTest(), WaveFunctionTester::runZeroVarianceTest(), sincos(), sMatrixElement(), SOECPComponent::sMatrixElements(), NESpaceGrid< REAL >::someMoreAxisGridStuff(), sph2cart(), kSpaceJastrow::StructureFactor(), TEST_CASE(), test_e2iphi(), YukawaBreakup< T >::Xk(), DerivYukawaBreakup< T >::Xk(), Ylm(), and Ylm().
|
inline |
Definition at line 1467 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
sincos function wrapper
Definition at line 62 of file math.hpp.
References cos(), qmcplusplus::Units::time::s, and sin().
Referenced by qmcplusplus::C2C::assign_v(), qmcplusplus::C2R::assign_v(), SplineC2R< ST >::assign_v(), SplineC2C< ST >::assign_v(), SplineC2COMPTarget< ST >::assign_v(), SplineC2ROMPTarget< ST >::assign_v(), SplineC2R< ST >::assign_vgh(), SplineC2C< ST >::assign_vgh(), SplineC2COMPTarget< ST >::assign_vgh(), SplineC2ROMPTarget< ST >::assign_vgh(), SplineC2R< ST >::assign_vghgh(), SplineC2C< ST >::assign_vghgh(), SplineC2COMPTarget< ST >::assign_vghgh(), SplineC2ROMPTarget< ST >::assign_vghgh(), qmcplusplus::C2C::assign_vgl(), qmcplusplus::C2R::assign_vgl(), SplineC2R< ST >::assign_vgl(), SplineC2C< ST >::assign_vgl(), SplineC2R< ST >::assign_vgl_from_l(), SplineC2C< ST >::assign_vgl_from_l(), SplineC2COMPTarget< ST >::assign_vgl_from_l(), SplineC2ROMPTarget< ST >::assign_vgl_from_l(), Gvectors< ST, LT >::calc_phase_shift(), compute_phase(), StructFact::computeRhok(), eval_e2iphi(), LCAOrbitalBuilder::EvalPeriodicImagePhaseFactors(), FreeOrbital::evaluate_notranspose(), Gvectors< ST, LT >::evaluate_psi_r(), SoaAtomicBasisSet< ROT, SH >::evaluateV(), FreeOrbital::evaluateValue(), SoaAtomicBasisSet< ROT, SH >::evaluateVGH(), SoaAtomicBasisSet< ROT, SH >::evaluateVGHGH(), FreeOrbital::evaluateVGL(), SoaAtomicBasisSet< ROT, SH >::evaluateVGL(), fix_phase_rotate(), fix_phase_rotate_c2r(), SoaAtomicBasisSet< ROT, SH >::mw_evaluateV(), SoaAtomicBasisSet< ROT, SH >::mw_evaluateVGL(), SpinorSet::mw_evaluateVGLandDetRatioGradsWithSpin(), StructFact::mw_updateAllPart(), and smoothing().
|
inline |
Definition at line 128 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by FnHypSin::operator()().
|
inline |
Definition at line 1475 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
| T smoothing | ( | smoothing_functions | func_id, |
| T | x, | ||
| T & | dx, | ||
| T & | d2x | ||
| ) |
1/2 - 1/2 tanh(alpha * (x - 1/2))
(1+cos(PI*(1-cos(PI*x))/2))/2
1-x
Definition at line 22 of file SmoothFunctions.cpp.
References BLAS::cone, COSCOS, LEKS2018, LINEAR, qmcplusplus::Units::time::s, sincos(), and tanh().
Referenced by HybridRepCenterOrbitals< SPLINEBASE::DataType >::selectRegionAndComputeSmoothing().
| template float smoothing | ( | smoothing_functions | func_id, |
| float | x, | ||
| float & | dx, | ||
| float & | d2x | ||
| ) |
| template double smoothing | ( | smoothing_functions | func_id, |
| double | x, | ||
| double & | dx, | ||
| double & | d2x | ||
| ) |
Definition at line 30 of file EinsplineSetBuilderESHDF.fft.cpp.
References BandInfo::BandIndex, qmcplusplus::Units::charge::e, BandInfo::Energy, and BandInfo::TwistIndex.
Referenced by EinsplineSetBuilder::OccupyBands_ESHDF().
| qmcplusplus::species_set | ( | iattribute | , |
| ispecies | |||
| ) |
|
inline |
wrapper for Ylm, which can take any normal position vector as an argument param[in] l angular momentum param[in] m magnetic quantum number param[in] r is a position vector.
This does not have to be normalized and is in the standard ordering [x,y,z]
Definition at line 141 of file Ylm.h.
References qmcplusplus::Units::distance::m, norm(), sqrt(), and Ylm().
Referenced by SOECPComponent::calculateProjector(), SOECPComponent::evaluateOneExactSpinIntegration(), SOECPComponent::evaluateValueAndDerivatives(), and TEST_CASE().
|
inline |
get cartesian derivatives of spherical Harmonics.
This takes a arbitrary position vector (x,y,z) and returns (d/dx, d/dy, d/dz)Ylm param[in] l angular momentum param[in] m magnetic quantum number param[in] r position vector. This does not have to be normalized and is in the standard ordering [x,y,z] param[out] grad (d/dx, d/dy, d/dz) of Ylm
Definition at line 158 of file Ylm.h.
References derivYlmSpherical(), qmcplusplus::Units::distance::m, norm(), and sqrt().
Referenced by TEST_CASE().
| qmcplusplus::SpinDensityNew | ( | std::move(sdi) | , |
| species_set | |||
| ) |
|
inline |
Definition at line 57 of file string_utils.h.
References qmcplusplus::Units::time::s, and whitespace().
Referenced by SpaceGrid::initialize_rectilinear(), ReferencePoints::put(), ReferencePointsInput::readRefPointsXML(), and TEST_CASE().
|
inline |
|
inline |
Split FFTs into real/imaginary components.
| in | ffts |
| out_r | real component |
| out_i | imaginary components |
Definition at line 193 of file einspline_helper.hpp.
References Array< T, D, ALLOC >::data().
Referenced by OneSplineOrbData::fft_spline().
| void splitPhiEta | ( | int | center, |
| const std::vector< bool > & | corrCenter, | ||
| LCAOrbitalSet & | Phi, | ||
| LCAOrbitalSet & | Eta | ||
| ) |
Divide molecular orbital into atomic S-orbitals on this center (phi), and everything else (eta).
Definition at line 226 of file CuspCorrectionConstruction.cpp.
References LCAOrbitalSet::C, LCAOrbitalSet::getBasisSetSize(), SPOSet::getOrbitalSetSize(), and LCAOrbitalSet::myBasisSet.
Referenced by applyCuspCorrection(), generateCuspInfo(), and TEST_CASE().
|
inline |
Definition at line 136 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by NESpaceGrid< REAL >::accumulate(), QMCCostFunctionBase::addCJParams(), SOVMCUpdateAll::advanceWalker(), SODMCUpdatePbyPWithRejectionFast::advanceWalker(), DMCUpdatePbyPWithRejectionFast::advanceWalker(), VMCUpdatePbyP::advanceWalker(), SOVMCUpdatePbyP::advanceWalker(), DMCUpdatePbyPL2::advanceWalker(), CSVMCUpdatePbyP::advanceWalker(), CSVMCUpdateAll::advanceWalker(), CSVMCUpdateAllWithDrift::advanceWalker(), RMCUpdatePbyPWithDrift::advanceWalkersRMC(), RMCUpdatePbyPWithDrift::advanceWalkersVMC(), SCTFunctor< SCT, 1 >::apply(), SCTFunctor< SCT, 2 >::apply(), DTD_BConds< T, D, SC >::apply_bc(), assignGaussRand(), AtomicOrbitals< ST >::AtomicOrbitals(), QMCFiniteSize::build_spherical_grid(), QMCFiniteSize::calcPotentialCorrection(), LatticeAnalyzer< T, 3 >::calcSimulationCellRadius(), LatticeAnalyzer< T, 2 >::calcSimulationCellRadius(), SpinDensityInput::calculateDerivedParameters(), MagnetizationDensityInput::calculateDerivedParameters(), LatticeAnalyzer< T, 3 >::calcWignerSeitzRadius(), LatticeAnalyzer< T, 2 >::calcWignerSeitzRadius(), cart2sph(), Cartesian2Spherical(), CartesianTensor< T, Point_t, Tensor_t, GGG_t >::CartesianTensor(), SpaceGrid::check_grid(), NESpaceGrid< REAL >::check_grid(), QMCUpdateBase::checkLogAndGL(), QMCDriverNew::checkLogAndGL(), Cholesky(), cholesky(), MPC::compute_g_G(), compute_norm(), compute_phase(), ACForce::compute_regularizer_f(), DTD_BConds< T, 3, SUPERCELL_OPEN+SOA_OFFSET >::computeDist(), DTD_BConds< T, 3, PPPO+SOA_OFFSET >::computeDist(), DTD_BConds< T, 3, PPPS+SOA_OFFSET >::computeDist(), DTD_BConds< T, 3, PPPG+SOA_OFFSET >::computeDist(), DTD_BConds< T, 3, PPNG+SOA_OFFSET >::computeDist(), DTD_BConds< T, 3, PPNO+SOA_OFFSET >::computeDist(), DTD_BConds< T, 3, PPNS+SOA_OFFSET >::computeDist(), DTD_BConds< T, 3, SUPERCELL_WIRE+SOA_OFFSET >::computeDist(), DTD_BConds< T, 3, SUPERCELL_OPEN+SOA_OFFSET >::computeDistances(), DTD_BConds< T, 3, PPPO+SOA_OFFSET >::computeDistances(), DTD_BConds< T, 3, PPPS+SOA_OFFSET >::computeDistances(), DTD_BConds< T, 3, PPPG+SOA_OFFSET >::computeDistances(), DTD_BConds< T, 3, PPNG+SOA_OFFSET >::computeDistances(), DTD_BConds< T, 3, PPNO+SOA_OFFSET >::computeDistances(), DTD_BConds< T, 3, PPNS+SOA_OFFSET >::computeDistances(), DTD_BConds< T, 3, SUPERCELL_WIRE+SOA_OFFSET >::computeDistances(), DTD_BConds< T, 3, SUPERCELL_OPEN+SOA_OFFSET >::computeDistancesOffload(), DTD_BConds< T, 3, PPPO+SOA_OFFSET >::computeDistancesOffload(), DTD_BConds< T, 3, PPPS+SOA_OFFSET >::computeDistancesOffload(), DTD_BConds< T, 3, PPPG+SOA_OFFSET >::computeDistancesOffload(), DTD_BConds< T, 3, PPNG+SOA_OFFSET >::computeDistancesOffload(), DTD_BConds< T, 3, PPNO+SOA_OFFSET >::computeDistancesOffload(), DTD_BConds< T, 3, PPNS+SOA_OFFSET >::computeDistancesOffload(), DTD_BConds< T, 3, SUPERCELL_WIRE+SOA_OFFSET >::computeDistancesOffload(), DescentEngine::computeFinalizationUncertainties(), RadialJastrowBuilder::computeJ2uk(), CountingJastrowBuilder::createCJ(), SHOSetBuilder::createSPOSet(), CubicFormula(), derivYlmSpherical(), DiracParser::DiracParser(), RPABFeeBreakup< T >::Dlindhard(), RPABFeeBreakup< T >::Dlindhardp(), dratioTWF(), Eig(), SpaceGrid::evaluate(), SphericalTensor< T, Point_t, Tensor_t, GGG_t >::evaluate(), SoaSphericalTensor< ST >::evaluate_bare(), SHOSet::evaluate_check(), SphericalTensor< T, Point_t, Tensor_t, GGG_t >::evaluateAll(), ExampleHeComponent::evaluateLog(), EwaldHandler2D::evaluateLR_r0(), EwaldHandlerQuasi2D::evaluateLR_r0(), EwaldHandler3D::evaluateLR_r0(), NonLocalECPComponent::evaluateOneBodyOpMatrixdRContribution(), NonLocalECPComponent::evaluateOneWithForces(), BackflowTransformation::evaluatePbyP(), BackflowTransformation::evaluatePbyPAll(), BackflowTransformation::evaluatePbyPWithGrad(), EwaldHandler2D::evaluateSR_k0(), SphericalTensor< T, Point_t, Tensor_t, GGG_t >::evaluateTest(), SoaAtomicBasisSet< ROT, SH >::evaluateV(), SoaAtomicBasisSet< ROT, SH >::evaluateVGH(), SoaAtomicBasisSet< ROT, SH >::evaluateVGHGH(), SoaAtomicBasisSet< ROT, SH >::evaluateVGL(), SoaSphericalTensor< ST >::evaluateVGL_impl(), SphericalTensor< T, Point_t, Tensor_t, GGG_t >::evaluateWithHessian(), EwaldHandler2D::EwaldHandler2D(), EwaldHandlerQuasi2D::EwaldHandlerQuasi2D(), qmcplusplus::ewaldref::ewaldSum(), EwaldHandler3D::filldFk_dk(), LRHandlerSRCoulomb< Func, BreakupBasis >::filldFk_dk(), EwaldHandler2D::fillFk(), EwaldHandlerQuasi2D::fillFk(), SkPot::FillFk(), LRHandlerTemp< Func, BreakupBasis >::fillFk(), LRHandlerSRCoulomb< Func, BreakupBasis >::fillYk(), LRHandlerSRCoulomb< Func, BreakupBasis >::fillYkg(), EwaldHandler3D::fillYkgstrain(), LRHandlerSRCoulomb< Func, BreakupBasis >::fillYkgstrain(), KContainer::findApproxMMax(), fix_phase_rotate_c2r(), found_shorter_base(), GaussianCombo< T >::GaussianCombo(), DensityMatrices1B::generate_density_samples(), DensityMatrices1B::generate_samples(), OneBodyDensityMatrices::generateDensitySamples(), OneBodyDensityMatrices::generateDensitySamplesWithSpin(), generateRotationMatrix(), OneBodyDensityMatrices::generateSamples(), DriftModifierUNR::getDrift(), getDriftScale(), qmcplusplus::ewaldref::getKappaEwald(), qmcplusplus::ewaldref::getKappaMadelung(), LinearMethod::getNonLinearRescale(), getScaledDrift(), getScaledDriftL2(), getStats(), Gvectors< ST, LT >::Gvectors(), MPC::init_f_G(), InitMolecularSystem::initAtom(), EwaldHandler3D::initBreakup(), SHOSet::initialize(), DensityMatrices1B::initialize(), J2KECorrection< RealType, FT >::J2KECorrection(), LegendrePll(), lMatrixElement(), SOECPComponent::lmMatrixElements(), EwaldHandler3D::lrDf(), qmcplusplus::ewaldref::madelungSum(), BackflowBuilder::makeLongRange_twoBody(), makeSphereRandom(), MomentumDistribution::MomentumDistribution(), DescentEngine::mpi_unbiased_ratio_of_means(), SoaAtomicBasisSet< ROT, SH >::mw_evaluateV(), SoaAtomicBasisSet< ROT, SH >::mw_evaluateVGL(), normalize(), DensityMatrices1B::normalize(), OneBodyDensityMatrices::normalizeBasis(), QMCFixedSampleLinearOptimizeBatched::one_shift_run(), OneBodyDensityMatrices::OneBodyDensityMatrices(), RspaceMadelungTerm::operator()(), STONorm< T >::operator()(), RPA0< T >::operator()(), RspaceEwaldTerm::operator()(), FnSqrt::operator()(), QMCCostFunctionBase::printCJParams(), WaveFunctionTester::printEloc(), QMCFiniteSize::printSkRawSphAvg(), GridExternalPotential::put(), StaticStructureFactor::put(), SpinDensity::put(), HarmonicExternalPotential::put(), MomentumEstimator::putSpecial(), Quadrature3D< T >::Quadrature3D(), ParticleSet::randomizeFromSource(), EshdfFile::readInEigFcn(), YukawaBreakup< T >::reset(), DerivYukawaBreakup< T >::reset(), CrystalLattice< ST, 3 >::reset(), QMCUpdateBase::resetRun(), WaveFunctionTester::runBasicTest(), DescentEngine::sample_finish(), SFNBranch::setBranchCutoff(), SimpleFixedNodeBranch::setBranchCutoff(), LRBreakupParameters< T, 3 >::SetLRCutoffs(), setScaledDriftPbyPandNodeCorr(), QMCUpdateBase::setTau(), kSpaceJastrow::setupGvecs(), LRBreakup< BreakupBasis >::SetupKVecs(), SimCellRad(), SkAllEstimator::SkAllEstimator(), SkEstimator::SkEstimator(), SkPot::SkPot(), EwaldHandlerQuasi2D::slab_vsr_k0(), SoaCartesianTensor< T >::SoaCartesianTensor(), SoaSphericalTensor< ST >::SoaSphericalTensor(), sphericalHarmonic(), sphericalHarmonicGrad(), SphericalTensor< T, Point_t, Tensor_t, GGG_t >::SphericalTensor(), EwaldHandler3D::srDf(), TauParams< RT, CoordsType::POS >::TauParams(), TauParams< RT, CoordsType::POS_SPIN >::TauParams(), TEST_CASE(), test_isnan(), TWF(), SHOSetBuilder::update_basis_states(), DescentEngine::updateParameters(), RPABFeeBreakup< T >::urpa(), DensityMatrices1B::warmup_sampling(), OneBodyDensityMatrices::warmupSampling(), WigSeitzRad(), RPABFeeBreakup< T >::Xk(), Ylm(), and Ylm().
|
inline |
Definition at line 1483 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
|
inline |
alternate to string2real calls c++ string to real conversion based on T's precision template<typename T>
Definition at line 74 of file ModernStringUtils.hpp.
|
inline |
Definition at line 115 of file string_utils.h.
References qmcplusplus::Units::time::s.
Referenced by SpaceGrid::initialize_rectilinear().
|
inline |
alternate to string2real calls c++ string to real conversion based on T's precision template<typename T>
Definition at line 52 of file ModernStringUtils.hpp.
|
inline |
Definition at line 117 of file string_utils.h.
References qmcplusplus::Units::time::s.
Referenced by SpaceGrid::initialize_rectilinear(), and ReferencePoints::put().
|
inline |
Definition at line 26 of file string_utils.h.
References qmcplusplus::Units::time::s.
Referenced by ReferencePoints::put(), ReferencePointsInput::readRefPointsXML(), and TEST_CASE().
|
inline |
Definition at line 170 of file ParticleAttribOps.h.
References Vector< T, Alloc >::size().
Referenced by BareKineticEnergy::evaluate(), BareKineticEnergy::evaluate_orig(), DiracDeterminantWithBackflow::evaluateDerivatives(), J1Spin< FT >::evaluateDerivatives(), TwoBodyJastrow< FT >::evaluateDerivatives(), J1OrbitalSoA< FT >::evaluateDerivatives(), BareKineticEnergy::evaluateWithIonDerivs(), WaveFunctionTester::runZeroVarianceTest(), TEST_CASE(), and test_J3_polynomial3D().
|
inline |
Definition at line 181 of file ParticleAttribOps.h.
| std::atomic<size_t> qmcplusplus::SYCLallocator_device_mem_allocated | ( | 0 | ) |
|
inline |
compute Trace(H*G)
gg is symmetrized as gg[0]=GG(0,0) gg[1]=GG(0,1)+GG(1,0) gg[2]=GG(0,2)+GG(2,0) gg[3]=GG(1,1) gg[4]=GG(1,2)+GG(2,1) gg[5]=GG(2,2)
Definition at line 45 of file contraction_helper.hpp.
Referenced by SplineC2R< ST >::assign_vgl(), SplineR2R< ST >::assign_vgl(), SplineC2C< ST >::assign_vgl(), SplineC2COMPTarget< ST >::evaluateVGL(), SplineC2ROMPTarget< ST >::evaluateVGL(), SplineC2COMPTarget< ST >::evaluateVGLMultiPos(), SplineC2ROMPTarget< ST >::evaluateVGLMultiPos(), SplineC2COMPTarget< ST >::mw_evaluateVGLandDetRatioGrads(), and SplineC2ROMPTarget< ST >::mw_evaluateVGLandDetRatioGrads().
| T qmcplusplus::t3_contract | ( | T | h000, |
| T | h001, | ||
| T | h002, | ||
| T | h011, | ||
| T | h012, | ||
| T | h022, | ||
| T | h111, | ||
| T | h112, | ||
| T | h122, | ||
| T | h222, | ||
| T | g1x, | ||
| T | g1y, | ||
| T | g1z, | ||
| T | g2x, | ||
| T | g2y, | ||
| T | g2z, | ||
| T | g3x, | ||
| T | g3y, | ||
| T | g3z | ||
| ) |
Coordinate transform for a 3rd rank symmetric tensor representing coordinate derivatives (hence t3_contract, for contraction with vectors).
hijk are the symmetry inequivalent tensor elements, i,j,k range from 0 to 2 for x to z. (gix,giy,giz) are vectors, labelled 1, 2 and 3. g1 is contracted with the first tensor index, g2 with the second, g3 with the third.
This would be easier with a for loop, but I'm sticking with the convention in this section.
Definition at line 69 of file contraction_helper.hpp.
Referenced by SplineC2R< ST >::assign_vghgh(), SplineR2R< ST >::assign_vghgh(), SplineC2C< ST >::assign_vghgh(), SplineC2COMPTarget< ST >::assign_vghgh(), and SplineC2ROMPTarget< ST >::assign_vghgh().
|
inline |
Definition at line 144 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by derivYlmSpherical(), and FnTan::operator()().
|
inline |
Definition at line 1491 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 152 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
Referenced by FnHypTan::operator()(), and smoothing().
|
inline |
Definition at line 1499 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
| qmcplusplus::TEMPLATE_TEST_CASE | ( | "RandomRotationMatrix" | , |
| "" | [numerics], | ||
| float | , | ||
| double | |||
| ) |
Definition at line 20 of file test_RotationMatrix3D.cpp.
References CHECK(), and qmcplusplus::Units::charge::e.
| qmcplusplus::TEMPLATE_TEST_CASE | ( | "cuSolverInverter" | , |
| "" | [wavefunction][fermion], | ||
| double | , | ||
| float | |||
| ) |
Definition at line 27 of file test_cuSolverInverter.cpp.
References CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), Matrix< T, Alloc >::cols(), Matrix< T, Alloc >::data(), fillIdentityMatrix(), TestMatrix1::fillInput(), TestMatrix1::fillInverse(), cuSolverInverter< T_FP >::invert_transpose(), TestMatrix1::logDet(), qmcplusplus::Units::distance::m, qmcplusplus::Units::force::N, Matrix< T, Alloc >::resize(), Matrix< T, Alloc >::rows(), and qmcplusplus::simd::transpose().
| qmcplusplus::TEMPLATE_TEST_CASE | ( | "rocSolverInverter" | , |
| "" | [wavefunction][fermion], | ||
| double | , | ||
| float | |||
| ) |
Definition at line 27 of file test_rocSolverInverter.cpp.
References CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), Matrix< T, Alloc >::cols(), Matrix< T, Alloc >::data(), fillIdentityMatrix(), TestMatrix1::fillInput(), TestMatrix1::fillInverse(), rocSolverInverter< T_FP >::invert_transpose(), TestMatrix1::logDet(), qmcplusplus::Units::distance::m, qmcplusplus::Units::force::N, Matrix< T, Alloc >::resize(), Matrix< T, Alloc >::rows(), and qmcplusplus::simd::transpose().
| qmcplusplus::TEMPLATE_TEST_CASE | ( | "ParseGridInput::Good" | , |
| "" | [estimators], | ||
| float | , | ||
| double | |||
| ) |
Definition at line 44 of file test_ParseGridInput.cpp.
References CHECK(), and qmcplusplus::Units::time::s.
| qmcplusplus::TEMPLATE_TEST_CASE | ( | "syclSolverInverter" | , |
| "" | [wavefunction][fermion], | ||
| double | , | ||
| float | |||
| ) |
Definition at line 52 of file test_syclSolverInverter.cpp.
| qmcplusplus::TEMPLATE_TEST_CASE | ( | "ParseGridInput::Bad" | , |
| "" | [estimators], | ||
| float | , | ||
| double | |||
| ) |
| qmcplusplus::TEMPLATE_TEST_CASE | ( | "ParseGridInput_constructors" | , |
| "" | [estimators], | ||
| float | , | ||
| double | |||
| ) |
Definition at line 147 of file test_ParseGridInput.cpp.
References CHECK().
| void qmcplusplus::test_C_diamond | ( | ) |
Definition at line 32 of file test_pyscf_complex_MO.cpp.
References CHECK(), OHMMS::Controller, SPOSetBuilderFactory::createSPOSetBuilder(), doc, qmcplusplus::Units::charge::e, Libxml2Document::getRoot(), Libxml2Document::getXPathContext(), imag(), lattice, okay, Libxml2Document::parse(), XMLParticleParser::readXML(), and REQUIRE().
Referenced by TEST_CASE().
| void qmcplusplus::test_cartesian_ao | ( | ) |
Definition at line 25 of file test_cartesian_ao.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), OHMMS::Controller, SPOSetBuilderFactory::createSPOSetBuilder(), doc, Libxml2Document::getRoot(), Libxml2Document::getXPathContext(), ispecies, okay, Libxml2Document::parse(), and REQUIRE().
Referenced by TEST_CASE().
| qmcplusplus::TEST_CASE | ( | "stdlib round" | , |
| "" | [numerics] | ||
| ) |
Definition at line 12 of file test_stdlib.cpp.
References REQUIRE().
| qmcplusplus::TEST_CASE | ( | "TWFGrads" | , |
| "" | [QMCWaveFunctions] | ||
| ) |
Definition at line 17 of file test_TWFGrads.cpp.
References CHECK(), POS, POS_SPIN, and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "StlPrettyPrint" | , |
| "" | [utilities] | ||
| ) |
Definition at line 18 of file test_StlPrettyPrint.cpp.
References CHECK().
| qmcplusplus::TEST_CASE | ( | "test_communicate_split_one" | , |
| "" | [message] | ||
| ) |
Definition at line 18 of file test_communciate.cpp.
References OHMMS::Controller, and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "SizeLimitedDataQueue" | , |
| "" | [estimators] | ||
| ) |
Definition at line 18 of file test_SizeLimitedDataQueue.cpp.
References CHECK(), SizeLimitedDataQueue< T, NUM_FIELDS >::push(), SizeLimitedDataQueue< T, NUM_FIELDS >::size(), and SizeLimitedDataQueue< T, NUM_FIELDS >::weighted_avg().
| qmcplusplus::TEST_CASE | ( | "Taus" | , |
| "" | [Particle] | ||
| ) |
Definition at line 18 of file test_TauParams.cpp.
References CHECK().
| qmcplusplus::TEST_CASE | ( | "NaNguard" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 19 of file test_NaNguard.cpp.
References NaNguard::checkOneParticleGradients(), NaNguard::checkOneParticleRatio(), and sqrt().
| qmcplusplus::TEST_CASE | ( | "Bessel" | , |
| "" | [numerics] | ||
| ) |
Definition at line 19 of file test_bessel.cpp.
References bessel_steed_array_cpu(), CHECK(), and qmcplusplus::Units::charge::e.
| qmcplusplus::TEST_CASE | ( | "test oneDimCubicSplineLinearGrid" | , |
| "" | [numerics] | ||
| ) |
Definition at line 19 of file test_OneDimCubicSplineLinearGrid.cpp.
References CHECK(), n, OneDimCubicSpline< Td, Tg, CTd, CTg >::spline(), and OneDimCubicSpline< Td, Tg, CTd, CTg >::splint().
| qmcplusplus::TEST_CASE | ( | "CheckSphericalIntegration" | , |
| "" | [numerics] | ||
| ) |
Definition at line 19 of file test_Quadrature.cpp.
References Quadrature3D< T >::quad_ok, and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "PlatformSelector" | , |
| "" | [platform] | ||
| ) |
Definition at line 19 of file test_PlatformSelector.cpp.
References CHECK(), CPU, CUDA, OMPTARGET, PlatformSelector< KIND >::selectPlatform(), and SYCL.
| qmcplusplus::TEST_CASE | ( | "DTModes" | , |
| "" | [particle] | ||
| ) |
Definition at line 19 of file test_DTModes.cpp.
References CHECK(), NEED_FULL_TABLE_ANYTIME, and NEED_TEMP_DATA_ON_HOST.
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A Ewald Quasi2D exception" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 19 of file test_ewald_quasi2d.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), ParticleSet::getSpeciesSet(), lattice, LRCoulombSingleton::QUASI2D, ParticleSet::R, ParticleSet::setName(), LRCoulombSingleton::this_lr_type, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "FakeRandom determinism" | , |
| "" | [utilities] | ||
| ) |
Definition at line 19 of file test_FakeRandom.cpp.
References CHECK().
| qmcplusplus::TEST_CASE | ( | "ci_configuration2" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 19 of file test_ci_configuration.cpp.
References ci_configuration2::calculateExcitations(), ci_configuration2::calculateNumOfExcitations(), CHECK(), REQUIRE(), and sign().
| qmcplusplus::TEST_CASE | ( | "string_utils_streamToVec" | , |
| "" | [utilities] | ||
| ) |
Definition at line 20 of file test_string_utils.cpp.
References CHECK(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "makeRefVector" | , |
| "" | [type_traits] | ||
| ) |
Definition at line 20 of file test_template_types.cpp.
References CHECK(), is_same(), and qmcplusplus::Units::time::s.
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A Ewald2D square" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 20 of file test_ewald2d.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAA::evaluate(), ParticleSet::getSpeciesSet(), lattice, ParticleSet::R, ParticleSet::setName(), LRCoulombSingleton::STRICT2D, LRCoulombSingleton::this_lr_type, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "EwaldRef" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 20 of file test_EwaldRef.cpp.
References qmcplusplus::Units::distance::A, CHECK(), and qmcplusplus::ewaldref::ewaldEnergy().
| qmcplusplus::TEST_CASE | ( | "NonLocalTOperator" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 20 of file test_NonLocalTOperator.cpp.
References CHECK(), NonLocalTOperator::groupByElectron(), REQUIRE(), NonLocalTOperator::selectMove(), and NonLocalTOperator::thingsThatShouldBeInMyConstructor().
| qmcplusplus::TEST_CASE | ( | "UtilityFunctions OpenMP" | , |
| "" | [concurrency] | ||
| ) |
Definition at line 20 of file test_UtilityFunctionsOPENMP.cpp.
References CHECK(), and omp_set_num_threads().
| qmcplusplus::TEST_CASE | ( | "ConstantSizeMatrix basics" | , |
| "" | [containers] | ||
| ) |
Definition at line 20 of file test_ConstantSizeMatrix.cpp.
References ConstantSizeMatrix< T, ALLOC >::capacity(), CHECK(), ConstantSizeMatrix< T, ALLOC >::resize(), and ConstantSizeMatrix< T, ALLOC >::size().
| qmcplusplus::TEST_CASE | ( | "RecordArray basics" | , |
| "" | [containers] | ||
| ) |
Definition at line 20 of file test_RecordArray.cpp.
References CHECK(), RecordArray< T >::getNumOfEntries(), RecordArray< T >::getNumOfParams(), REQUIRE(), and RecordArray< T >::resize().
| qmcplusplus::TEST_CASE | ( | "kcontainer at gamma in 3D" | , |
| "" | [longrange] | ||
| ) |
Definition at line 20 of file test_kcontainer.cpp.
References CHECK(), qmcplusplus::Units::charge::e, KContainer::kpts, KContainer::kpts_cart, lattice, sqrt(), and KContainer::updateKLists().
| qmcplusplus::TEST_CASE | ( | "Aligned allocator" | , |
| "" | [numerics] | ||
| ) |
Definition at line 21 of file test_aligned_allocator.cpp.
References REQUIRE().
| qmcplusplus::TEST_CASE | ( | "OMP runtime memory" | , |
| "" | [OMP] | ||
| ) |
Definition at line 21 of file test_runtime_mem.cpp.
References print_mem().
| qmcplusplus::TEST_CASE | ( | "SpaceGridInputs::parseXML::valid" | , |
| "" | [estimators] | ||
| ) |
Definition at line 21 of file test_SpaceGridInput.cpp.
References doc, Libxml2Document::getRoot(), makeSpaceGridInput(), node, okay, Libxml2Document::parseFromString(), and ValidSpinDensityInput::xml.
| qmcplusplus::TEST_CASE | ( | "VMCDriverInput readXML" | , |
| "" | [drivers] | ||
| ) |
Definition at line 21 of file test_VMCDriverInput.cpp.
References doc, VMCDriverInput::get_use_drift(), Libxml2Document::getRoot(), node, okay, Libxml2Document::parseFromString(), VMCDriverInput::readXML(), REQUIRE(), qmcplusplus::testing::valid_vmc_input_sections, and qmcplusplus::testing::valid_vmc_input_vmc_batch_index.
| qmcplusplus::TEST_CASE | ( | "MCCoords" | , |
| "" | [Particle] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "ModernStringUtils_strToLower" | , |
| "" | [utilities] | ||
| ) |
Definition at line 21 of file test_ModernStringUtils.cpp.
References CHECK(), and lowerCase().
| qmcplusplus::TEST_CASE | ( | "ConstantSPOSet" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 21 of file test_ConstantSPOSet.cpp.
References CHECK(), CHECKED_ELSE(), checkMatrix(), ParticleSet::create(), OHMMS_DIM, and Matrix< T, Alloc >::resize().
| qmcplusplus::TEST_CASE | ( | "ShortRangeCuspJastrowFunctor" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 22 of file test_short_range_cusp_jastrow.cpp.
References ShortRangeCuspFunctor< T >::A, ShortRangeCuspFunctor< T >::B, CHECK(), ShortRangeCuspFunctor< T >::checkInVariablesExclusive(), OHMMS::Controller, OptimizableFunctorBase::cutoff_radius, doc, ShortRangeCuspFunctor< T >::evaluate(), ShortRangeCuspFunctor< T >::evaluateDerivatives(), Libxml2Document::getRoot(), ShortRangeCuspFunctor< T >::ID_A, ShortRangeCuspFunctor< T >::ID_B, ShortRangeCuspFunctor< T >::ID_R0, VariableSet::name(), ShortRangeCuspFunctor< T >::Opt_A, ShortRangeCuspFunctor< T >::Opt_B, ShortRangeCuspFunctor< T >::Opt_R0, Libxml2Document::parseFromString(), ShortRangeCuspFunctor< T >::put(), ShortRangeCuspFunctor< T >::R0, REQUIRE(), VariableSet::resetIndex(), ShortRangeCuspFunctor< T >::resetParametersExclusive(), and VariableSet::size_of_active().
| qmcplusplus::TEST_CASE | ( | "UserJastrowFunctor" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 22 of file test_user_jastrow.cpp.
References UserFunctor< T >::A, UserFunctor< T >::B, CHECK(), UserFunctor< T >::checkInVariablesExclusive(), OHMMS::Controller, doc, UserFunctor< T >::evaluate(), UserFunctor< T >::evaluateDerivatives(), Libxml2Document::getRoot(), VariableSet::name(), okay, Libxml2Document::parseFromString(), UserFunctor< T >::put(), REQUIRE(), VariableSet::resetIndex(), UserFunctor< T >::resetParametersExclusive(), UserFunctor< T >::setCusp(), and VariableSet::size_of_active().
| qmcplusplus::TEST_CASE | ( | "OMPmath" | , |
| "" | [OMP] | ||
| ) |
Definition at line 22 of file test_math.cpp.
References qmcplusplus::Units::distance::A, CHECK(), cos(), qmcplusplus::Units::time::s, and sin().
| qmcplusplus::TEST_CASE | ( | "SYCL_allocator" | , |
| "" | [SYCL] | ||
| ) |
Definition at line 22 of file test_SYCLallocator.cpp.
References CHECK(), Vector< T, Alloc >::data(), getSYCLDefaultDeviceDefaultQueue(), and queue.
| qmcplusplus::TEST_CASE | ( | "WFOptDriverInput Instantiation" | , |
| "" | [drivers] | ||
| ) |
Definition at line 22 of file test_WFOptDriverInput.cpp.
| qmcplusplus::TEST_CASE | ( | "QMCDriverInput Instantiation" | , |
| "" | [drivers] | ||
| ) |
Definition at line 22 of file test_QMCDriverInput.cpp.
Definition at line 22 of file test_drift.cpp.
References CHECK(), ParticleSet::create(), ParticleSet::G, ParticleSet::setName(), setScaledDriftPbyPandNodeCorr(), and sqrt().
| qmcplusplus::TEST_CASE | ( | "Hybrid Engine query store" | , |
| "" | [drivers][hybrid] | ||
| ) |
This provides a basic test of the hybrid engine's check on whether vectors need to be stored.
Definition at line 22 of file test_HybridEngine.cpp.
| qmcplusplus::TEST_CASE | ( | "VirtualParticleSet" | , |
| "" | [particle] | ||
| ) |
Definition at line 22 of file test_VirtualParticleSet.cpp.
References CHECK(), OHMMS::Controller, VirtualParticleSet::createResource(), DistanceTableAB::getDistances(), ParticleSet::getDistTableAB(), MinimalParticlePool::make_NiO_a4(), VirtualParticleSet::makeMoves(), ParticleSet::R, REQUIRE(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "J1 evaluate derivatives Jastrow" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 22 of file test_J1OrbitalSoA.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), ParticleSet::addTable(), app_log(), WaveFunctionFactory::buildTWF(), CHECK(), OHMMS::Controller, ParticleSet::create(), doc, ParticleSet::get(), ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), ispecies, okay, Libxml2Document::parseFromString(), ParticleSet::R, VariableSet::removeInactive(), REQUIRE(), ParticleSet::resetGroups(), ParticleSet::setName(), VariableSet::size_of_active(), twf, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "StdRandom mt19937 determinism" | , |
| "" | [utilities] | ||
| ) |
Definition at line 23 of file test_StdRandom.cpp.
References CHECK().
| qmcplusplus::TEST_CASE | ( | "Scalar Estimator Input" | , |
| "" | [estimators] | ||
| ) |
Definition at line 23 of file test_ScalarEstimatorInputs.cpp.
References CHECK(), doc, LocalEnergyInput::get_type(), CSLocalEnergyInput::get_type(), RMCLocalEnergyInput::get_type(), Libxml2Document::getRoot(), getXMLAttributeValue(), lowerCase(), node, okay, Libxml2Document::parseFromString(), REQUIRE(), and ValidSpinDensityInput::xml.
| qmcplusplus::TEST_CASE | ( | "TwoBodyJastrow simple" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 23 of file test_J2_derivatives.cpp.
References TwoBodyJastrow< FT >::checkOutVariables(), ParticleSet::create(), and ParticleSet::setName().
| qmcplusplus::TEST_CASE | ( | "spline_function_1" | , |
| "" | [numerics] | ||
| ) |
Definition at line 23 of file test_one_dim_cubic_spline.cpp.
References CHECK(), CubicSplineSolve(), qmcplusplus::Units::charge::e, and n.
| qmcplusplus::TEST_CASE | ( | "solveGeneralizedEigenvalues" | , |
| "" | [drivers] | ||
| ) |
Definition at line 23 of file test_Eigensolver.cpp.
References app_log(), CHECK(), qmcplusplus::Units::force::N, and Eigensolver::solveGeneralizedEigenvalues().
| qmcplusplus::TEST_CASE | ( | "WaveFunctionFactory" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 23 of file test_wavefunction_factory.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), WaveFunctionFactory::buildTWF(), OHMMS::Controller, doc, Libxml2Document::getRoot(), okay, Libxml2Document::parseFromString(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "CUDA_allocators" | , |
| "" | [CUDA] | ||
| ) |
Definition at line 23 of file test_CUDAallocator.cpp.
References cudaErrorCheck(), cudaMemoryTypeDevice, cudaMemoryTypeHost, cudaMemoryTypeManaged, cudaPointerAttributes, cudaPointerGetAttributes, Vector< T, Alloc >::data(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "CompositeSPO::diamond_1x1x1" | , |
| "[wavefunction" | |||
| ) |
Definition at line 23 of file test_CompositeSPOSet.cpp.
References CompositeSPOSet::add(), ProjectData::BATCH, CHECK(), comm, OHMMS::Controller, doc, CompositeSPOSet::evaluate_notranspose(), SPOSet::getOrbitalSetSize(), ProjectData::getRuntimeOptions(), MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), outputManager, particle_pool, OutputManagerClass::pause(), pset, SPOSet::size(), test_project, twf, and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "SoaDistanceTableAA compute_size" | , |
| "" | [distance_table] | ||
| ) |
Definition at line 23 of file test_SoaDistanceTableAA.cpp.
References CHECK(), SoaDistanceTableAA< T, D, SC >::compute_size(), ParticleSet::create(), ParticleSet::getTotalNum(), and ParticleSet::setName().
| qmcplusplus::TEST_CASE | ( | "VMCFactory Instantiation" | , |
| "" | [drivers] | ||
| ) |
Definition at line 23 of file test_VMCFactoryNew.cpp.
References doc, Libxml2Document::getRoot(), node, okay, Libxml2Document::parseFromString(), REQUIRE(), UPDATE_MODE, qmcplusplus::testing::valid_vmc_input_sections, and qmcplusplus::testing::valid_vmc_input_vmc_batch_index.
| qmcplusplus::TEST_CASE | ( | "selectEigenvalues" | , |
| "" | [drivers] | ||
| ) |
Definition at line 23 of file test_LinearMethod.cpp.
References CHECK(), and LinearMethod::selectEigenvalue().
| qmcplusplus::TEST_CASE | ( | "make_seed" | , |
| "" | [utilities] | ||
| ) |
Definition at line 24 of file test_rng.cpp.
References make_seed(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "compute_batch_parameters" | , |
| "" | [drivers] | ||
| ) |
Definition at line 24 of file test_QMCCostFunctionBatched.cpp.
References batch_size, CHECK(), and compute_batch_parameters().
| qmcplusplus::TEST_CASE | ( | "Density Estimator" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 24 of file test_density_estimator.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), ParticleSet::create(), DensityEstimator::evaluate(), ParticleSet::getSpeciesSet(), ParticleSet::R, ParticleSet::resetGroups(), OperatorBase::setHistories(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "WFOptDriverInput readXML" | , |
| "" | [drivers] | ||
| ) |
Definition at line 24 of file test_WFOptDriverInput.cpp.
References doc, WFOptDriverInput::get_opt_crowd_size(), WFOptDriverInput::get_opt_method(), WFOptDriverInput::get_opt_num_crowds(), Libxml2Document::getRoot(), node, okay, Libxml2Document::parseFromString(), WFOptDriverInput::readXML(), REQUIRE(), and qmcplusplus::testing::valid_opt_input_sections.
| qmcplusplus::TEST_CASE | ( | "Legendre" | , |
| "" | [numerics] | ||
| ) |
Definition at line 24 of file test_ylm.cpp.
References abs(), CHECK(), and LegendrePll().
| qmcplusplus::TEST_CASE | ( | "StructFact" | , |
| "" | [lrhandler] | ||
| ) |
evalaute bare Coulomb in 3D using LRHandlerTemp
Definition at line 24 of file test_StructFact.cpp.
References SpeciesSet::addSpecies(), app_log(), CrystalLattice< T, D >::BoxBConds, CHECK(), ParticleSet::create(), Tensor< T, D >::diagonal(), qmcplusplus::Units::charge::e, SimulationCell::getKLists(), ParticleSet::getLRBox(), ParticleSet::getSpeciesSet(), ParticleSet::groups(), KContainer::numk, ParticleSet::R, CrystalLattice< T, D >::R, REQUIRE(), CrystalLattice< T, D >::reset(), CrystalLattice< T, D >::Rv, StructFact::updateAllPart(), and CrystalLattice< T, D >::Volume.
| qmcplusplus::TEST_CASE | ( | "array" | , |
| "" | [OhmmsPETE] | ||
| ) |
Definition at line 24 of file test_Array.cpp.
References qmcplusplus::Units::distance::A, B(), CHECK(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "matrix" | , |
| "" | [OhmmsPETE] | ||
| ) |
Definition at line 24 of file test_Matrix.cpp.
References qmcplusplus::Units::distance::A, B(), qmcplusplus::Units::charge::C, CHECK(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "ParallelExecutor<STD> function case" | , |
| "" | [concurrency] | ||
| ) |
Definition at line 24 of file test_ParallelExecutorSTD.cpp.
References REQUIRE(), and TestTask().
| qmcplusplus::TEST_CASE | ( | "QMCDriverInput readXML" | , |
| "" | [drivers] | ||
| ) |
Definition at line 24 of file test_QMCDriverInput.cpp.
References doc, QMCDriverInput::get_qmc_method(), Libxml2Document::getRoot(), node, okay, Libxml2Document::parseFromString(), QMCDriverInput::readXML(), REQUIRE(), qmcplusplus::testing::valid_dmc_input_dmc_batch_index, qmcplusplus::testing::valid_dmc_input_sections, qmcplusplus::testing::valid_vmc_input_sections, and qmcplusplus::testing::valid_vmc_input_vmc_batch_index.
| qmcplusplus::TEST_CASE | ( | "ParticleSet distance table management" | , |
| "" | [particle] | ||
| ) |
Definition at line 24 of file test_ParticleSet.cpp.
References ParticleSet::addTable(), ParticleSet::getDistTable(), REQUIRE(), and ParticleSet::setName().
| qmcplusplus::TEST_CASE | ( | "cuSolverInverter_bench" | , |
| "" | [wavefunction][benchmark] | ||
| ) |
Definition at line 24 of file benchmark_cuSolverInverter.cpp.
References check_matrix_result, CHECKED_ELSE(), checkMatrix(), cuSolverInverter< T_FP >::invert_transpose(), DiracMatrix< T_FP >::invert_transpose(), qmcplusplus::Units::distance::m, qmcplusplus::testing::makeRngSpdMatrix(), qmcplusplus::Units::force::N, and Matrix< T, Alloc >::resize().
| qmcplusplus::TEST_CASE | ( | "LogGridLight" | , |
| "" | [wavefunction][LCAO] | ||
| ) |
Definition at line 25 of file test_multiquintic_spline.cpp.
References CHECK(), LogGridLight< T >::getCLForQuintic(), LogGridLight< T >::locate(), REQUIRE(), and LogGridLight< T >::set().
| qmcplusplus::TEST_CASE | ( | "ReferencePointsInput::parseXML::valid" | , |
| "" | [estimators] | ||
| ) |
Definition at line 25 of file test_ReferencePointsInput.cpp.
References doc, Libxml2Document::getRoot(), node, okay, and Libxml2Document::parseFromString().
| qmcplusplus::TEST_CASE | ( | "ewald3d" | , |
| "" | [lrhandler] | ||
| ) |
evalaute bare Coulomb using EwaldHandler3D
Definition at line 25 of file test_ewald3d.cpp.
References CrystalLattice< T, D >::BoxBConds, CHECK(), ParticleSet::createSK(), Tensor< T, D >::diagonal(), EwaldHandler3D::evaluate(), EwaldHandler3D::evaluateLR(), EwaldHandler3D::initBreakup(), LRHandlerBase::LR_kc, LRHandlerBase::LR_rc, LRHandlerBase::MaxKshell, CrystalLattice< T, D >::R, CrystalLattice< T, D >::reset(), CrystalLattice< T, D >::Rv, EwaldHandler3D::Sigma, sqrt(), and CrystalLattice< T, D >::Volume.
| qmcplusplus::TEST_CASE | ( | "vector" | , |
| "" | [OhmmsPETE] | ||
| ) |
Definition at line 25 of file test_Vector.cpp.
References qmcplusplus::Units::distance::A, B(), qmcplusplus::Units::charge::C, CHECK(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "vector" | , |
| "" | [OhmmsSoA] | ||
| ) |
Definition at line 25 of file test_vector_soa.cpp.
References CHECK().
| qmcplusplus::TEST_CASE | ( | "Basic Gaussian" | , |
| "" | [numerics] | ||
| ) |
Definition at line 25 of file test_gaussian_basis.cpp.
References CHECK(), GaussianCombo< T >::BasicGaussian::df(), GaussianCombo< T >::BasicGaussian::evaluate(), GaussianCombo< T >::BasicGaussian::f(), and GaussianCombo< T >::BasicGaussian::reset().
| qmcplusplus::TEST_CASE | ( | "parsewords_empty" | , |
| "" | [utilities] | ||
| ) |
Definition at line 25 of file test_parser.cpp.
References parsewords(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "prime number set 32 bit" | , |
| "" | [utilities] | ||
| ) |
Definition at line 26 of file test_prime_set.cpp.
References PrimeNumberSet< UIntType >::get(), REQUIRE(), and PrimeNumberSet< UIntType >::size().
| qmcplusplus::TEST_CASE | ( | "ParallelExecutor<OPENMP> function case" | , |
| "" | [concurrency] | ||
| ) |
Definition at line 26 of file test_ParallelExecutorOPENMP.cpp.
References omp_get_max_threads(), REQUIRE(), and TestTaskOMP().
| qmcplusplus::TEST_CASE | ( | "SpinDensityInput::readXML" | , |
| "" | [estimators] | ||
| ) |
Definition at line 26 of file test_SpinDensityInput.cpp.
References SpinDensityInput::calculateDerivedParameters(), CHECK(), doc, CrystalLattice< T, D >::explicitly_defined, SpinDensityInput::DerivedParameters::gdims, SpinDensityInput::get_cell(), Libxml2Document::getRoot(), SpinDensityInput::DerivedParameters::grid, lattice, qmcplusplus::testing::makeTestLattice(), node, SpinDensityInput::DerivedParameters::npoints, okay, Libxml2Document::parseFromString(), REQUIRE(), sdi, and ValidSpinDensityInput::xml.
| qmcplusplus::TEST_CASE | ( | "LRBreakupParameters" | , |
| "" | [lattice] | ||
| ) |
Lattice is defined but Open BC is also used.
Definition at line 26 of file test_LRBreakupParameters.cpp.
References CHECK().
| qmcplusplus::TEST_CASE | ( | "FS parse Sk file" | , |
| "" | [tools] | ||
| ) |
Definition at line 26 of file test_qmcfstool.cpp.
References CHECK().
| qmcplusplus::TEST_CASE | ( | "SampleStack" | , |
| "" | [particle] | ||
| ) |
Definition at line 26 of file test_sample_stack.cpp.
References SampleStack::appendSample(), CHECK(), SampleStack::clearEnsemble(), MCSample::convertToWalker(), SampleStack::getGlobalNumSamples(), SampleStack::getMaxSamples(), SampleStack::getNumSamples(), SampleStack::getSample(), REQUIRE(), SampleStack::resetSampleCount(), and SampleStack::setMaxSamples().
| qmcplusplus::TEST_CASE | ( | "rocSolverInverter_bench" | , |
| "" | [wavefunction][benchmark] | ||
| ) |
Definition at line 26 of file benchmark_rocSolverInverter.cpp.
References check_matrix_result, CHECKED_ELSE(), checkMatrix(), rocSolverInverter< T_FP >::invert_transpose(), DiracMatrix< T_FP >::invert_transpose(), qmcplusplus::Units::distance::m, qmcplusplus::testing::makeRngSpdMatrix(), qmcplusplus::Units::force::N, and Matrix< T, Alloc >::resize().
| qmcplusplus::TEST_CASE | ( | "Resource" | , |
| "" | [utilities] | ||
| ) |
Definition at line 27 of file test_ResourceCollection.cpp.
References REQUIRE().
| qmcplusplus::TEST_CASE | ( | "J1 spin evaluate derivatives Jastrow" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 27 of file test_J1Spin.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), OHMMS::Controller, ParticleSet::create(), doc, ParticleSet::G, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), ispecies, ParticleSet::L, log(), okay, Libxml2Document::parseFromString(), ParticleSet::R, VariableSet::removeInactive(), REQUIRE(), ParticleSet::resetGroups(), ParticleSet::setName(), VariableSet::size_of_active(), twf, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "QMCTypes" | , |
| "" | [type_traits] | ||
| ) |
Definition at line 27 of file test_qmctypes.cpp.
| qmcplusplus::TEST_CASE | ( | "EngineHandle construction" | , |
| "" | [drivers] | ||
| ) |
This provides a basic test of constructing an EngineHandle object and checking information in it.
Definition at line 27 of file test_EngineHandle.cpp.
References app_log(), OHMMS::Controller, doc, Libxml2Document::getRoot(), okay, Libxml2Document::parseFromString(), DescentEngine::processXML(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "OneBodyDensityMatricesInput::from_xml" | , |
| "" | [estimators] | ||
| ) |
Definition at line 27 of file test_OneBodyDensityMatricesInput.cpp.
References doc, Libxml2Document::getRoot(), node, obdmi, okay, Libxml2Document::parseFromString(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "checkMatrix_OhmmsMatrix_real" | , |
| "" | [utilities][for_testing] | ||
| ) |
Definition at line 27 of file test_checkMatrix.cpp.
References CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), qmcplusplus::Units::charge::e, REQUIRE(), and Matrix< T, Alloc >::resize().
| qmcplusplus::TEST_CASE | ( | "FakeRandom set_value" | , |
| "" | [utilities] | ||
| ) |
Definition at line 27 of file test_FakeRandom.cpp.
References CHECK(), and FakeRandom< T >::set_value().
| qmcplusplus::TEST_CASE | ( | "RandomNumberControl make_seeds" | , |
| "" | [ohmmsapp] | ||
| ) |
Definition at line 28 of file test_rng_control.cpp.
References RandomNumberControl::Children, RandomNumberControl::make_seeds(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "Crystal_lattice_periodic_bulk" | , |
| "" | [lattice] | ||
| ) |
Lattice is defined but Open BC is also used.
Definition at line 28 of file test_CrystalLattice.cpp.
References CrystalLattice< T, D >::BoxBConds, CHECK(), Tensor< T, D >::diagonal(), CrystalLattice< T, D >::isValid(), CrystalLattice< T, D >::outOfBound(), CrystalLattice< T, D >::R, REQUIRE(), CrystalLattice< T, D >::reset(), and CrystalLattice< T, D >::Volume.
| qmcplusplus::TEST_CASE | ( | "Cartesian Tensor" | , |
| "" | [numerics] | ||
| ) |
Definition at line 28 of file test_cartesian_tensor.cpp.
References CHECK(), CartesianTensor< T, Point_t, Tensor_t, GGG_t >::evaluate(), and CartesianTensor< T, Point_t, Tensor_t, GGG_t >::getYlm().
| qmcplusplus::TEST_CASE | ( | "double_1d_grid_functor" | , |
| "" | [numerics] | ||
| ) |
Definition at line 28 of file test_grid_functor.cpp.
References CHECK(), OneDimGridBase< T, CT >::dh(), OneDimGridBase< T, CT >::dr(), LinearGrid< T, CT >::makeClone(), REQUIRE(), OneDimGridBase< T, CT >::rmax(), OneDimGridBase< T, CT >::rmin(), LinearGrid< T, CT >::set(), and OneDimGridBase< T, CT >::size().
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A Ewald3D" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 28 of file test_coulomb_pbcAA_ewald.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAA::evalConsts(), CoulombPBCAA::evaluate(), ParticleSet::getSpeciesSet(), lattice, ParticleSet::R, and ParticleSet::setName().
| qmcplusplus::TEST_CASE | ( | "MCPopulation::createWalkers" | , |
| "" | [particle][population] | ||
| ) |
Definition at line 28 of file test_MCPopulation.cpp.
References CHECK(), comm, OHMMS::Controller, WalkerConfigurations::createWalkers(), MCPopulation::createWalkers(), WalkerConfigurations::getActiveWalkers(), hamiltonian_pool, MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), MinimalHamiltonianPool::make_hamWithEE(), particle_pool, Communicate::rank(), WalkerConfigurations::resize(), twf, and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "particle_attrib_scalar" | , |
| "" | [particle_base] | ||
| ) |
Definition at line 28 of file test_particle_attrib.cpp.
References REQUIRE(), Vector< T, Alloc >::resize(), and Vector< T, Alloc >::size().
| qmcplusplus::TEST_CASE | ( | "read_lattice_xml" | , |
| "" | [particle_io][xml] | ||
| ) |
Definition at line 28 of file test_lattice_parser.cpp.
References CHECK(), doc, Libxml2Document::getRoot(), okay, Libxml2Document::parseFromString(), LatticeParser::put(), CrystalLattice< T, D >::R, REQUIRE(), and CrystalLattice< T, D >::Volume.
| qmcplusplus::TEST_CASE | ( | "read_particleset_xml" | , |
| "" | [particle_io][xml] | ||
| ) |
Definition at line 28 of file test_xml_io.cpp.
References CHECK(), doc, OhmmsElementBase::getName(), Libxml2Document::getRoot(), ParticleSet::groups(), okay, Libxml2Document::parseFromString(), ParticleSet::R, XMLParticleParser::readXML(), REQUIRE(), and Vector< T, Alloc >::size().
| qmcplusplus::TEST_CASE | ( | "read_particle_mass_same_xml" | , |
| "" | [particle_io][xml] | ||
| ) |
Definition at line 28 of file test_xml_mass.cpp.
References SpeciesSet::addAttribute(), CHECK(), doc, OhmmsElementBase::getName(), Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), ParticleSet::getTotalNum(), ParticleSet::GroupID, ParticleSet::isSameMass(), okay, Libxml2Document::parseFromString(), XMLParticleParser::readXML(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "MagnetizationDensityInput::from_xml" | , |
| "" | [estimators] | ||
| ) |
Definition at line 28 of file test_MagnetizationDensityInput.cpp.
References app_log(), MagnetizationDensityInput::calculateDerivedParameters(), CHECK(), doc, MagnetizationDensityInput::get_corner(), MagnetizationDensityInput::get_dr(), MagnetizationDensityInput::get_grid(), MagnetizationDensityInput::get_integrator(), MagnetizationDensityInput::get_nsamples(), Libxml2Document::getRoot(), qmcplusplus::testing::invalid_mag_density_input_sections, lattice, qmcplusplus::testing::makeTestLattice(), node, okay, Libxml2Document::parseFromString(), REQUIRE(), qmcplusplus::testing::magdensity::valid_mag_density_input_sections, and qmcplusplus::testing::magdensity::valid_magdensity_input.
| qmcplusplus::TEST_CASE | ( | "ParticleSetPool" | , |
| "" | [qmcapp] | ||
| ) |
Definition at line 29 of file test_particle_pool.cpp.
References ParticleSetPool::addParticleSet(), OHMMS::Controller, doc, ParticleSetPool::get(), ParticleSetPool::getParticleSet(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSetPool::getWalkerSet(), okay, Libxml2Document::parseFromString(), ParticleSetPool::put(), ParticleSetPool::randomize(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "PlaneWave SPO from HDF for BCC H" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 29 of file test_pw.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), OHMMS::Controller, ParticleSet::create(), SPOSetBuilder::createSPOSet(), doc, Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), imag(), lattice, okay, Libxml2Document::parseFromString(), ParticleSet::R, real(), REQUIRE(), ParticleSet::resetGroups(), ParticleSet::setName(), ParticleSetPool::setSimulationCell(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "ProjectData" | , |
| "" | [ohmmsapp] | ||
| ) |
Definition at line 29 of file test_project_data.cpp.
References OHMMS::Controller, ProjectData::getTitle(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-B Ewald3D" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 29 of file test_coulomb_pbcAB_ewald.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAB::evalConsts(), CoulombPBCAA::evaluate(), CoulombPBCAB::evaluate(), ParticleSet::getSpeciesSet(), lattice, ParticleSet::R, ParticleSet::resetGroups(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "WalkerLogCollector::collect" | , |
| "" | [estimators] | ||
| ) |
Definition at line 29 of file test_WalkerLogCollector.cpp.
References app_log(), ProjectData::BATCH, CHECK(), WalkerLogCollector::collect(), comm, OHMMS::Controller, ProjectData::getRuntimeOptions(), ham, hamiltonian_pool, MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), MinimalHamiltonianPool::make_hamWithEE(), qmcplusplus::hdf::num_walkers, particle_pool, pset, test_project, twf, walker, WalkerLogCollector::walker_particle_real_buffer, WalkerLogCollector::walker_property_int_buffer, WalkerLogCollector::walker_property_real_buffer, qmcplusplus::hdf::walkers, and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "OMPdeepcopy" | , |
| "" | [OMP] | ||
| ) |
Definition at line 29 of file test_deep_copy.cpp.
References OMPallocator< T, HostAllocator >::allocate(), OMPallocator< T, HostAllocator >::deallocate(), REQUIRE(), and container::size.
| qmcplusplus::TEST_CASE | ( | "EstimatorManagerBase" | , |
| "" | [estimators] | ||
| ) |
Definition at line 29 of file test_manager.cpp.
References EstimatorManagerBase::add(), OHMMS::Controller, EstimatorManagerBase::getEstimator(), REQUIRE(), EstimatorManagerBase::reset(), EstimatorManagerBase::size(), EstimatorManagerBase::start(), and EstimatorManagerBase::stop().
| qmcplusplus::TEST_CASE | ( | "DescentEngine RMSprop update" | , |
| "" | [drivers][descent] | ||
| ) |
This provides a basic test of the descent engine's parameter update algorithm.
Definition at line 29 of file test_DescentEngine.cpp.
References app_log(), CHECK(), OHMMS::Controller, doc, Libxml2Document::getRoot(), mean(), n, okay, Libxml2Document::parseFromString(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "QMCDriverNew tiny case" | , |
| "" | [drivers] | ||
| ) |
Definition at line 29 of file test_QMCDriverNew.cpp.
References ProjectData::BATCH, comm, OHMMS::Controller, doc, QMCDriverNew::get_num_living_walkers(), QMCDriverInterface::getBranchEngine(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), hamiltonian_pool, MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), MinimalHamiltonianPool::make_hamWithEE(), node, okay, outputManager, Libxml2Document::parseFromString(), particle_pool, OutputManagerClass::pause(), QMCDriverNewTestWrapper::process(), Communicate::rank(), QMCDriverInput::readXML(), REQUIRE(), OutputManagerClass::resume(), QMCDriverNew::setStatus(), Communicate::size(), test_project, qmcplusplus::testing::valid_vmc_input_sections, qmcplusplus::testing::valid_vmc_input_vmc_tiny_index, and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "MPIExceptionWrapper function case" | , |
| "" | [Utilities] | ||
| ) |
Definition at line 29 of file test_mpi_exception_wrapper.cpp.
References comm, OHMMS::Controller, and mpiTestFunctionWrapped().
| qmcplusplus::TEST_CASE | ( | "open_bconds" | , |
| "" | [lattice] | ||
| ) |
Definition at line 30 of file test_ParticleBConds.cpp.
References DTD_BConds< T, D, SC >::apply_bc(), CHECK(), DTD_BConds< T, D, SC >::evaluate_rsquared(), OHMMS_PRECISION, and sqrt().
| qmcplusplus::TEST_CASE | ( | "ExampleHe" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 30 of file test_example_he.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ExampleHeComponent::B, WaveFunctionFactory::buildTWF(), CHECK(), ExampleHeComponent::checkInVariablesExclusive(), ExampleHeComponent::checkOutVariables(), OHMMS::Controller, doc, Dot(), ExampleHeComponent::evalGrad(), ExampleHeComponent::evaluateDerivatives(), ExampleHeComponent::evaluateLog(), Libxml2Document::getRoot(), imag(), okay, Libxml2Document::parseFromString(), ExampleHeComponent::ratio(), ExampleHeComponent::ratioGrad(), REQUIRE(), ExampleHeComponent::resetParametersExclusive(), Vector< T, Alloc >::resize(), VariableSet::size_of_active(), and Sum().
| qmcplusplus::TEST_CASE | ( | "readCuspInfo" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 30 of file test_soa_cusp_corr.cpp.
References qmcplusplus::Units::charge::C, CHECK(), OHMMS::Controller, okay, readCuspInfo(), REQUIRE(), and Matrix< T, Alloc >::resize().
| qmcplusplus::TEST_CASE | ( | "dummy" | , |
| "" | [lrhandler] | ||
| ) |
evalaute bare Coulomb using DummyLRHandler
Definition at line 30 of file test_lrhandler.cpp.
References CrystalLattice< T, D >::BoxBConds, CHECK(), ParticleSet::createSK(), Tensor< T, D >::diagonal(), LRHandlerBase::Fk_symm, ParticleSet::getSimulationCell(), DummyLRHandler< Func >::initBreakup(), LRHandlerBase::LR_kc, LRHandlerBase::LR_rc, LRHandlerBase::MaxKshell, norm(), CrystalLattice< T, D >::R, CrystalLattice< T, D >::reset(), CrystalLattice< T, D >::Rv, and CrystalLattice< T, D >::Volume.
| qmcplusplus::TEST_CASE | ( | "Jastrow 2D" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 30 of file test_2d_jastrow.cpp.
References ParticleSet::addTable(), app_log(), RadialJastrowBuilder::buildComponent(), CHECK(), OHMMS::Controller, doc, ParticleSet::G, Libxml2Document::getRoot(), ParticleSet::getTotalNum(), imag(), ParticleSet::L, lattice, qmcplusplus::Units::distance::m, ParticleSet::makeMove(), ParticleSet::makeVirtualMoves(), node, OHMMS_DIM, okay, Libxml2Document::parseFromString(), ParticleSet::R, XMLParticleParser::readXML(), real(), ParticleSet::rejectMove(), REQUIRE(), Vector< T, Alloc >::resize(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "Gaussian Functor" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 30 of file test_counting_jastrow.cpp.
References CHECK(), CountingGaussian::evaluate(), CountingGaussian::evaluateDerivatives(), CountingGaussian::evaluateLog(), CountingGaussian::evaluateLogDerivatives(), Libxml2Document::getRoot(), Libxml2Document::parseFromString(), CountingGaussian::put(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "distance_open_z" | , |
| "" | [distance_table][xml] | ||
| ) |
Definition at line 30 of file test_distance_table.cpp.
References ParticleSet::addTable(), CHECK(), doc, ParticleSet::getDistTableAB(), OhmmsElementBase::getName(), Libxml2Document::getRoot(), ParticleSet::getTotalNum(), ParticleSet::isSameMass(), okay, Libxml2Document::parseFromString(), XMLParticleParser::readXML(), REQUIRE(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "kspace jastrow" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 31 of file test_kspace_jastrow.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), app_log(), kSpaceJastrowBuilder::buildComponent(), CHECK(), OHMMS::Controller, ParticleSet::create(), ParticleSet::createSK(), doc, ParticleSet::G, Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), ParticleSet::L, lattice, okay, Libxml2Document::parseFromString(), LatticeParser::put(), ParticleSet::R, REQUIRE(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "RPA Jastrow" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 31 of file test_rpa_jastrow.cpp.
References SpeciesSet::addSpecies(), app_log(), CHECK(), ParticleSet::create(), ParticleSet::createSK(), doc, ParticleSet::G, Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), ParticleSet::L, lattice, okay, Libxml2Document::parseFromString(), LatticeParser::put(), ParticleSet::R, REQUIRE(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "Stress BCC H Ewald3D" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 31 of file test_stress.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), LRCoulombSingleton::CoulombDerivHandler, LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), StressPBC::evaluate(), ParticleSet::getSpeciesSet(), ForceBase::getStressEE(), ForceBase::getStressEI(), lattice, ParticleSet::R, ParticleSet::resetGroups(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "LocalEnergyOnly" | , |
| "" | [estimators] | ||
| ) |
Definition at line 31 of file test_local_energy_est.cpp.
References LocalEnergyOnlyEstimator::accumulate(), WalkerConfigurations::begin(), CHECK(), ParticleSet::create(), MCWalkerConfiguration::createWalkers(), WalkerConfigurations::end(), LocalEnergyOnlyEstimator::getName(), ScalarEstimatorBase::scalars, and ParticleSet::setName().
| qmcplusplus::TEST_CASE | ( | "test_runtime_manager" | , |
| "" | [utilities] | ||
| ) |
Definition at line 32 of file test_runtime_manager.cpp.
References CHECK(), convert_to_ns(), qmcplusplus::Units::charge::e, RunTimeManager< CLOCK >::elapsed(), and qmcplusplus::Units::time::s.
| qmcplusplus::TEST_CASE | ( | "Einspline SPO from HDF diamond_1x1x1" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 32 of file test_einset.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), CHECK(), OHMMS::Controller, ParticleSet::create(), ParticleSet::createResource(), EinsplineSetBuilder::createSPOSetFromXML(), doc, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), lattice, okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), ParticleSet::setName(), ParticleSetPool::setSimulationCell(), Vector< T, Alloc >::size(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "LCAOrbitalBuilder" | , |
| "" | [wavefunction][LCAO] | ||
| ) |
Definition at line 32 of file test_LCAOrbitalBuilder.cpp.
References SpeciesSet::addSpecies(), CHECK(), OHMMS::Controller, ParticleSet::create(), doc, LCAOrbitalBuilder::getBasissetMap(), Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), okay, Libxml2Document::parseFromString(), REQUIRE(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "Spline applyRotation zero rotation" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 32 of file test_spline_applyrotation.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), CHECK(), CHECKED_ELSE(), checkMatrix(), OHMMS::Controller, ParticleSet::create(), EinsplineSetBuilder::createSPOSetFromXML(), doc, qmcplusplus::Units::charge::e, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), lattice, norm(), okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), ParticleSet::setName(), ParticleSetPool::setSimulationCell(), Vector< T, Alloc >::size(), and sqrt().
| qmcplusplus::TEST_CASE | ( | "SoA Cartesian Tensor" | , |
| "" | [numerics] | ||
| ) |
Definition at line 32 of file test_soa_cartesian_tensor.cpp.
References CHECK(), SoaCartesianTensor< T >::evaluateV(), and SoaCartesianTensor< T >::getcXYZ().
| qmcplusplus::TEST_CASE | ( | "EstimatorManagerCrowd::EstimatorManagerCrowd" | , |
| "" | [estimators] | ||
| ) |
Definition at line 32 of file test_EstimatorManagerCrowd.cpp.
References ProjectData::BATCH, CHECK(), comm, OHMMS::Controller, qmcplusplus::testing::createEstimatorManagerNewInputXML(), emi(), emn, estimators_doc, EstimatorManagerNew::getNumEstimators(), EstimatorManagerNew::getNumScalarEstimators(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), ham, hamiltonian_pool, MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), MinimalHamiltonianPool::make_hamWithEE(), particle_pool, pset, test_project, twf, and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "isnan" | , |
| "" | [numerics] | ||
| ) |
Definition at line 32 of file test_math.cpp.
| qmcplusplus::TEST_CASE | ( | "TraceManager" | , |
| "" | [estimators] | ||
| ) |
Definition at line 32 of file test_trace_manager.cpp.
References TraceManager::checkout_int(), TraceRequest::contribute_scalar(), OHMMS::Controller, TraceRequest::determine_stream_write(), TraceRequest::incorporate(), TraceManager::put(), TraceManager::request, TraceRequest::streaming_scalar(), and TraceManager::update_status().
| qmcplusplus::TEST_CASE | ( | "StdRandom save and load" | , |
| "" | [utilities] | ||
| ) |
Definition at line 33 of file test_StdRandom.cpp.
References CHECK(), and StdRandom< T >::init().
| qmcplusplus::TEST_CASE | ( | "Pade functor" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 33 of file test_pade_jastrow.cpp.
References qmcplusplus::Units::distance::A, B(), PadeFunctor< T >::B0, CHECK(), PadeFunctor< T >::evaluate(), and PadeFunctor< T >::setCusp().
| qmcplusplus::TEST_CASE | ( | "ParallelExecutor<STD> lambda case" | , |
| "" | [concurrency] | ||
| ) |
Definition at line 33 of file test_ParallelExecutorSTD.cpp.
References REQUIRE().
| qmcplusplus::TEST_CASE | ( | "HamiltonianPool" | , |
| "" | [qmcapp] | ||
| ) |
Definition at line 33 of file test_hamiltonian_pool.cpp.
References WaveFunctionPool::addFactory(), ParticleSetPool::addParticleSet(), WaveFunctionFactory::buildEmptyTWFForTesting(), OHMMS::Controller, createElectronParticleSet(), doc, HamiltonianPool::empty(), HamiltonianPool::getHamiltonian(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), okay, Libxml2Document::parseFromString(), HamiltonianPool::put(), REQUIRE(), and QMCHamiltonian::size().
| qmcplusplus::TEST_CASE | ( | "pack scalar" | , |
| "" | [utilities] | ||
| ) |
Definition at line 33 of file test_pooled_memory.cpp.
References PooledMemory< T_scalar, Alloc >::add(), PooledMemory< T_scalar, Alloc >::allocate(), PooledData< T >::byteSize(), PooledMemory< T_scalar, Alloc >::byteSize(), PooledMemory< T_scalar, Alloc >::Current, PooledMemory< T_scalar, Alloc >::current(), PooledMemory< T_scalar, Alloc >::data(), Timer::elapsed(), PooledMemory< T_scalar, Alloc >::get(), PooledMemory< T_scalar, Alloc >::put(), REQUIRE(), PooledData< T >::resize(), Timer::restart(), and PooledMemory< T_scalar, Alloc >::rewind().
| qmcplusplus::TEST_CASE | ( | "Chiesa Force BCC H Ewald3D" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 33 of file test_force_ewald.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), OHMMS::Controller, LRCoulombSingleton::CoulombDerivHandler, LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), ForceChiesaPBCAA::evaluate(), ForceBase::getForces(), ForceBase::getForcesIonIon(), ParticleSet::getSpeciesSet(), ForceChiesaPBCAA::InitMatrix(), lattice, ParticleSet::R, ParticleSet::resetGroups(), ForceBase::setAddIonIon(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "Einspline SPO from HDF NiO a16 97 electrons" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 33 of file test_einset_NiO_a16.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), app_log(), CHECK(), OHMMS::Controller, ParticleSet::create(), ParticleSet::createResource(), EinsplineSetBuilder::createSPOSetFromXML(), Vector< T, Alloc >::device_data(), QMCTraits::DIM_VGL, doc, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), imag(), ispecies, Lattice, lattice, okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), Array< T, D, ALLOC >::resize(), ParticleSet::setName(), ParticleSetPool::setSimulationCell(), Vector< T, Alloc >::size(), and Vector< T, Alloc >::updateTo().
| qmcplusplus::TEST_CASE | ( | "lattice gaussian" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 34 of file test_lattice_gaussian.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), app_log(), LatticeGaussianProductBuilder::buildComponent(), CHECK(), OHMMS::Controller, doc, Dot(), exp(), Libxml2Document::getRoot(), lattice, log(), okay, Libxml2Document::parseFromString(), LatticeParser::put(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "QMCHamiltonian::flex_evaluate" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 34 of file test_QMCHamiltonian.cpp.
References comm, OHMMS::Controller, hamiltonian_pool, MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), MinimalHamiltonianPool::make_hamWithEE(), particle_pool, twf, and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "Fixed node branch" | , |
| "" | [drivers][walker_control] | ||
| ) |
Definition at line 34 of file test_fixed_node_branch.cpp.
References SimpleFixedNodeBranch::advanceQMCCounter(), SimpleFixedNodeBranch::B_COUNTER, CHECK(), OHMMS::Controller, SimpleFixedNodeBranch::getEstimatorManager(), SimpleFixedNodeBranch::getTau(), SimpleFixedNodeBranch::iParam, REQUIRE(), and SimpleFixedNodeBranch::setEstimatorManager().
| qmcplusplus::TEST_CASE | ( | "ObservableHelper::set_dimensions" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 34 of file test_ObservableHelper.cpp.
References CHECK(), dims, ObservableHelper::lower_bound, oh, and ObservableHelper::set_dimensions().
| qmcplusplus::TEST_CASE | ( | "Hybridrep SPO from HDF diamond_1x1x1" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 34 of file test_hybridrep.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), CHECK(), CHECKED_ELSE(), checkMatrix(), OHMMS::Controller, ParticleSet::create(), EinsplineSetBuilder::createSPOSetFromXML(), doc, qmcplusplus::Units::charge::e, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), lattice, okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), ParticleSet::setName(), ParticleSetPool::setSimulationCell(), Vector< T, Alloc >::size(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "walker" | , |
| "" | [particle] | ||
| ) |
Definition at line 34 of file test_walker.cpp.
References CHECK(), Walker< t_traits, p_traits >::R, and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "FairDivideLow_one" | , |
| "" | [utilities] | ||
| ) |
Definition at line 34 of file test_partition.cpp.
References FairDivideLow(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "ParallelExecutor<OPENMP> lambda case" | , |
| "" | [concurrency] | ||
| ) |
Definition at line 35 of file test_ParallelExecutorOPENMP.cpp.
References omp_get_max_threads(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "RandomNumberControl no random in xml" | , |
| "" | [ohmmsapp] | ||
| ) |
Definition at line 35 of file test_rng_control.cpp.
References doc, Libxml2Document::getXPathContext(), RandomNumberControl::initialize(), okay, Libxml2Document::parseFromString(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "TwoBodyJastrow one species and two variables" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 35 of file test_J2_derivatives.cpp.
References TwoBodyJastrow< FT >::addFunc(), CHECK(), TwoBodyJastrow< FT >::checkOutVariables(), ParticleSet::create(), VariableSet::insertFrom(), ParticleSet::setName(), and VariableSet::size_of_active().
| qmcplusplus::TEST_CASE | ( | "Bare Force" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 35 of file test_force.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), OHMMS::Controller, ParticleSet::create(), BareForce::evaluate(), ForceBase::getForces(), ParticleSet::getSpeciesSet(), ParticleSet::R, ParticleSet::resetGroups(), ForceBase::setAddIonIon(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "gaussian random array length 1" | , |
| "" | [particle_base] | ||
| ) |
Definition at line 35 of file test_random_seq.cpp.
References assignGaussRand(), and CHECK().
| qmcplusplus::TEST_CASE | ( | "DiracMatrix_identity" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 36 of file test_DiracMatrix.cpp.
References CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), fillIdentityMatrix(), DiracMatrix< T_FP >::invert_transpose(), qmcplusplus::Units::distance::m, and Matrix< T, Alloc >::resize().
| qmcplusplus::TEST_CASE | ( | "Bare Kinetic Energy" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 36 of file test_bare_kinetic.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), ParticleSet::create(), doc, BareKineticEnergy::evaluate(), ParticleSet::G, Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), ParticleSet::L, okay, Libxml2Document::parseFromString(), BareKineticEnergy::put(), ParticleSet::R, REQUIRE(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "ModernStringUtils_split" | , |
| "" | [utilities] | ||
| ) |
Definition at line 36 of file test_ModernStringUtils.cpp.
References CHECK(), qmcplusplus::modernstrutil::split(), and split().
| qmcplusplus::TEST_CASE | ( | "transform2gridfunctor" | , |
| "" | [numerics] | ||
| ) |
Definition at line 36 of file test_transform.cpp.
References CHECK(), and OneDimQuinticSpline< Td, Tg, CTd, CTg >::set().
| qmcplusplus::TEST_CASE | ( | "srcoul" | , |
| "" | [lrhandler] | ||
| ) |
evalaute bare Coulomb in 3D
Definition at line 36 of file test_srcoul.cpp.
References CrystalLattice< T, D >::BoxBConds, CHECK(), ParticleSet::createSK(), Tensor< T, D >::diagonal(), LRHandlerSRCoulomb< Func, BreakupBasis >::evaluate(), LRHandlerSRCoulomb< Func, BreakupBasis >::initBreakup(), LRHandlerBase::LR_kc, LRHandlerBase::LR_rc, LRHandlerBase::MaxKshell, CrystalLattice< T, D >::R, CrystalLattice< T, D >::reset(), CrystalLattice< T, D >::Rv, and CrystalLattice< T, D >::Volume.
| qmcplusplus::TEST_CASE | ( | "FakeRandom read write" | , |
| "" | [utilities] | ||
| ) |
Definition at line 36 of file test_FakeRandom.cpp.
References CHECK(), and FakeRandom< T >::set_value().
| qmcplusplus::TEST_CASE | ( | "DummyResource" | , |
| "" | [utilities] | ||
| ) |
Definition at line 37 of file test_ResourceCollection.cpp.
References DummyResource::makeClone(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "DiracMatrixComputeCUDA_cuBLAS_geam_call" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 37 of file test_DiracMatrixComputeCUDA.cpp.
References qmcplusplus::Units::distance::A, qmcplusplus::syclBLAS::copy_n(), CUBLAS_OP_N, CUBLAS_OP_T, cublasErrorCheck, cudaCheck, cudaMemcpyAsync, cudaMemcpyHostToDevice, Matrix< T, Alloc >::data(), Matrix< T, Alloc >::device_data(), qmcplusplus::cuBLAS::geam(), BLASHandle< PlatformKind::CUDA >::h_cublas, lda, n, queue, Matrix< T, Alloc >::resize(), and Matrix< T, Alloc >::size().
| qmcplusplus::TEST_CASE | ( | "PolynomialFunctor3D functor zero" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 37 of file test_polynomial_eeI_jastrow.cpp.
References PolynomialFunctor3D::evaluate(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-B" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 37 of file test_coulomb_pbcAB.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAA::evaluate(), CoulombPBCAB::evaluate(), ParticleSet::getSpeciesSet(), lattice, CoulombPBCAB::myConst, ParticleSet::R, ParticleSet::resetGroups(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "temp3d" | , |
| "" | [lrhandler] | ||
| ) |
evalaute bare Coulomb in 3D using LRHandlerTemp
Definition at line 37 of file test_temp.cpp.
References CrystalLattice< T, D >::BoxBConds, CHECK(), ParticleSet::createSK(), Tensor< T, D >::diagonal(), LRHandlerTemp< Func, BreakupBasis >::evaluate(), LRHandlerTemp< Func, BreakupBasis >::evaluateLR(), LRHandlerTemp< Func, BreakupBasis >::initBreakup(), LRHandlerBase::LR_kc, LRHandlerBase::LR_rc, LRHandlerBase::MaxKshell, CrystalLattice< T, D >::R, CrystalLattice< T, D >::reset(), CrystalLattice< T, D >::Rv, and CrystalLattice< T, D >::Volume.
| qmcplusplus::TEST_CASE | ( | "SpaceGridInputs::parseXML::invalid" | , |
| "" | [estimators] | ||
| ) |
Definition at line 37 of file test_SpaceGridInput.cpp.
References doc, Libxml2Document::getRoot(), node, okay, Libxml2Document::parseFromString(), REQUIRE(), and ValidSpinDensityInput::xml.
| qmcplusplus::TEST_CASE | ( | "BSpline builder Jastrow J2" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 38 of file test_J2_bspline.cpp.
References ParticleSet::acceptMove(), SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), app_log(), RadialJastrowBuilder::buildComponent(), CHECK(), OHMMS::Controller, ParticleSet::create(), BsplineFunctor< REAL >::CuspValue, BsplineFunctor< REAL >::DeltaR, doc, Dot(), BsplineFunctor< REAL >::evaluate(), ParticleSet::G, Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), ParticleSet::getTotalNum(), ParticleSet::L, qmcplusplus::Units::distance::m, ParticleSet::makeMove(), VirtualParticleSet::makeMoves(), ParticleSet::makeVirtualMoves(), qmcplusplus::Units::force::N, n, BsplineFunctor< REAL >::NumParams, OHMMS_DIM, okay, Libxml2Document::parseFromString(), ParticleSet::R, ParticleSet::rejectMove(), REQUIRE(), ParticleSet::resetGroups(), Vector< T, Alloc >::resize(), ParticleSet::setName(), Sum(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "complex_helper" | , |
| "" | [type_traits] | ||
| ) |
Definition at line 38 of file test_complex_helper.cpp.
References TestComplexHelper< P >::run().
| qmcplusplus::TEST_CASE | ( | "SpaceWarp" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 38 of file test_spacewarp.cpp.
References ParticleSet::addTable(), app_log(), CHECK(), SpaceWarpTransformation::computeSWT(), OHMMS::Controller, SpaceWarpTransformation::df(), doc, SpaceWarpTransformation::f(), ParticleSet::G, Libxml2Document::getRoot(), ParticleSet::getTotalNum(), Libxml2Document::getXPathContext(), ParticleSet::groups(), okay, Libxml2Document::parse(), ParticleSet::R, XMLParticleParser::readXML(), REQUIRE(), SpaceWarpTransformation::setPow(), Vector< T, Alloc >::size(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "SYCL_host_allocator" | , |
| "" | [SYCL] | ||
| ) |
Definition at line 39 of file test_SYCLallocator.cpp.
References CHECK(), Vector< T, Alloc >::data(), getSYCLDefaultDeviceDefaultQueue(), and queue.
| qmcplusplus::TEST_CASE | ( | "BSpline functor zero" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 39 of file test_J1_bspline.cpp.
References BsplineFunctor< REAL >::evaluate(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "spline_function_2" | , |
| "" | [numerics] | ||
| ) |
Definition at line 39 of file test_one_dim_cubic_spline.cpp.
References CHECK(), CubicSplineSolve(), qmcplusplus::Units::charge::e, and n.
| qmcplusplus::TEST_CASE | ( | "ReferencePointsInput::parseXML::invalid" | , |
| "" | [estimators] | ||
| ) |
Definition at line 39 of file test_ReferencePointsInput.cpp.
References doc, Libxml2Document::getRoot(), node, okay, Libxml2Document::parseFromString(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "DiracMatrixComputeOMPTarget_different_batch_sizes" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 40 of file test_DiracMatrixComputeOMPTarget.cpp.
References qmcplusplus::Units::distance::A, CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), qmcplusplus::syclBLAS::copy_n(), Matrix< T, Alloc >::data(), DiracMatrixComputeOMPTarget< VALUE_FP >::invert_transpose(), log_values(), DiracMatrixComputeOMPTarget< VALUE_FP >::mw_invertTranspose(), queue, and Matrix< T, Alloc >::resize().
| qmcplusplus::TEST_CASE | ( | "RotatedSPOs via SplineR2R" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 40 of file test_RotatedSPOs.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), CHECK(), OHMMS::Controller, ParticleSet::create(), EinsplineSetBuilder::createSPOSetFromXML(), doc, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), lattice, okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), ParticleSet::setName(), ParticleSetPool::setSimulationCell(), and Vector< T, Alloc >::size().
| qmcplusplus::TEST_CASE | ( | "Spherical Harmonics" | , |
| "" | [numerics] | ||
| ) |
Definition at line 40 of file test_ylm.cpp.
References CHECK(), and Ylm().
| qmcplusplus::TEST_CASE | ( | "SkAll" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 40 of file test_SkAllEstimator.cpp.
References SkAllEstimator::addObservables(), ParticleSet::addTable(), app_log(), ParticleSet::Collectables, OHMMS::Controller, doc, SkAllEstimator::evaluate(), SkAllEstimator::get(), ParticleSet::get(), ParticleSet::getDistTable(), OhmmsElementBase::getName(), ParticleSetPool::getParticleSet(), Libxml2Document::getRoot(), ParticleSet::getSimulationCell(), ParticleSet::isSameMass(), lat_xml, Libxml2Document::parseFromString(), ParticleSet::PropertyList, SkAllEstimator::put(), ParticleSetPool::put(), ParticleSet::R, ParticleSetPool::readSimulationCellXML(), REQUIRE(), OperatorBase::setHistories(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "MPIExceptionWrapper lambda case" | , |
| "" | [Utilities] | ||
| ) |
Definition at line 40 of file test_mpi_exception_wrapper.cpp.
References CHECK(), comm, OHMMS::Controller, and Communicate::size().
| qmcplusplus::TEST_CASE | ( | "VMC Particle-by-Particle advanceWalkers" | , |
| "" | [drivers][vmc] | ||
| ) |
Definition at line 41 of file test_vmc.cpp.
References SpeciesSet::addAttribute(), TrialWaveFunction::addComponent(), QMCHamiltonian::addObservables(), QMCHamiltonian::addOperator(), SpeciesSet::addSpecies(), ParticleSet::addTable(), QMCUpdateBase::advanceWalkers(), WalkerConfigurations::begin(), CHECK(), ParticleSet::create(), MCWalkerConfiguration::createWalkers(), WalkerConfigurations::end(), ParticleSet::getSpeciesSet(), QMCUpdateBase::nAccept, QMCUpdateBase::nReject, ParticleSet::R, TrialWaveFunction::registerData(), REQUIRE(), QMCUpdateBase::resetRun(), MCWalkerConfiguration::resetWalkerProperty(), ParticleSet::setName(), QMCUpdateBase::startBlock(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 41 of file test_coulomb_pbcAA.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAA::evalConsts(), CoulombPBCAA::evaluate(), CoulombPBCAA::get_madelung_constant(), ParticleSet::getSpeciesSet(), lattice, ParticleSet::R, and ParticleSet::setName().
| qmcplusplus::TEST_CASE | ( | "DMC Particle-by-Particle advanceWalkers ConstantOrbital" | , |
| "" | [drivers][dmc] | ||
| ) |
Definition at line 41 of file test_dmc.cpp.
References SpeciesSet::addAttribute(), TrialWaveFunction::addComponent(), QMCHamiltonian::addObservables(), QMCHamiltonian::addOperator(), SpeciesSet::addSpecies(), ParticleSet::addTable(), QMCUpdateBase::advanceWalkers(), WalkerConfigurations::begin(), CHECK(), ParticleSet::create(), MCWalkerConfiguration::createWalkers(), WalkerConfigurations::end(), ParticleSet::getSpeciesSet(), QMCUpdateBase::nAccept, QMCUpdateBase::nReject, ParticleSet::R, TrialWaveFunction::registerData(), REQUIRE(), QMCUpdateBase::resetRun(), MCWalkerConfiguration::resetWalkerProperty(), ParticleSet::setName(), QMCUpdateBase::startBlock(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "VMC" | , |
| "" | [drivers][vmc] | ||
| ) |
Definition at line 41 of file test_vmc_driver.cpp.
References CloneManager::acceptRatio(), SpeciesSet::addAttribute(), TrialWaveFunction::addComponent(), QMCHamiltonian::addOperator(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), CloneManager::clearClones(), OHMMS::Controller, ParticleSet::create(), MCWalkerConfiguration::createWalkers(), doc, Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), ParticleSet::getSpeciesSet(), okay, omp_get_max_threads(), Libxml2Document::parseFromString(), QMCDriver::process(), ParticleSet::R, REQUIRE(), MCWalkerConfiguration::resetWalkerProperty(), VMC::run(), Communicate::setName(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "double_1d_grid_functor_vs_n" | , |
| "" | [numerics] | ||
| ) |
Definition at line 42 of file test_grid_functor.cpp.
References CHECK(), OneDimGridBase< T, CT >::dh(), OneDimGridBase< T, CT >::dr(), n, REQUIRE(), OneDimGridBase< T, CT >::rmax(), OneDimGridBase< T, CT >::rmin(), LinearGrid< T, CT >::set(), and OneDimGridBase< T, CT >::size().
| qmcplusplus::TEST_CASE | ( | "EstimatorManagerInput::testInserts" | , |
| "" | [estimators] | ||
| ) |
Definition at line 42 of file test_EstimatorManagerInput.cpp.
References doc, emi(), Libxml2Document::getRoot(), node, okay, Libxml2Document::parseFromString(), REQUIRE(), EstimatorManagerInputTests::testAppendFromXML(), and ValidSpinDensityInput::xml.
| qmcplusplus::TEST_CASE | ( | "test_loop_timer" | , |
| "" | [utilities] | ||
| ) |
Definition at line 42 of file test_runtime_manager.cpp.
References CHECK(), convert_to_ns(), LoopTimer< CLOCK >::get_time_per_iteration(), qmcplusplus::Units::time::s, LoopTimer< CLOCK >::start(), and LoopTimer< CLOCK >::stop().
| qmcplusplus::TEST_CASE | ( | "DMC" | , |
| "" | [drivers][dmc] | ||
| ) |
Definition at line 42 of file test_dmc_driver.cpp.
References CloneManager::acceptRatio(), SpeciesSet::addAttribute(), TrialWaveFunction::addComponent(), QMCHamiltonian::addOperator(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), CloneManager::clearClones(), OHMMS::Controller, ParticleSet::create(), MCWalkerConfiguration::createWalkers(), doc, Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), ParticleSet::getSpeciesSet(), okay, omp_get_max_threads(), Libxml2Document::parseFromString(), QMCDriver::process(), ParticleSet::R, REQUIRE(), DMC::run(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "TrialWaveFunction_diamondC_1x1x1" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 43 of file test_TrialWaveFunction.cpp.
References TrialWaveFunction::acceptMove(), SpeciesSet::addAttribute(), TrialWaveFunction::addComponent(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), ParticleSet::addTable(), app_log(), RadialJastrowBuilder::buildComponent(), TrialWaveFunction::calcRatio(), CHECK(), OHMMS::Controller, ParticleSet::create(), TrialWaveFunction::createResource(), ParticleSet::createSK(), EinsplineSetBuilder::createSPOSetFromXML(), DC_POS, DC_POS_OFFLOAD, doc, qmcplusplus::Units::charge::e, TrialWaveFunction::evaluateLog(), TrialWaveFunction::extractOptimizableObjectRefs(), TrialWaveFunction::FERMIONIC, TrialWaveFunction::findMSD(), TrialWaveFunction::getLogPsi(), ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), TWFGrads< CoordsType::POS >::grads_positions, lattice, TrialWaveFunction::makeClone(), TrialWaveFunction::mw_accept_rejectMove(), TrialWaveFunction::mw_calcRatio(), TrialWaveFunction::mw_calcRatioGrad(), TrialWaveFunction::mw_evalGrad(), TrialWaveFunction::mw_evaluateLog(), ParticleSet::mw_makeMove(), ParticleSet::mw_update(), TrialWaveFunction::NONFERMIONIC, okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), ParticleSet::resetGroups(), ParticleSet::setName(), ParticleSetPool::setSimulationCell(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "e2iphi" | , |
| "" | [numerics] | ||
| ) |
Definition at line 43 of file test_e2iphi.cpp.
| qmcplusplus::TEST_CASE | ( | "ParallelExecutor<STD> nested case" | , |
| "" | [concurrency] | ||
| ) |
Definition at line 43 of file test_ParallelExecutorSTD.cpp.
References REQUIRE(), and TestTask().
| qmcplusplus::TEST_CASE | ( | "OutputManager basic" | , |
| "" | [utilities] | ||
| ) |
Definition at line 43 of file test_output_manager.cpp.
References DEBUG, HIGH, OutputManagerClass::isDebugActive(), OutputManagerClass::isHighActive(), LOW, REQUIRE(), and OutputManagerClass::setVerbosity().
| qmcplusplus::TEST_CASE | ( | "FairDivideLow_two" | , |
| "" | [utilities] | ||
| ) |
Definition at line 43 of file test_partition.cpp.
References FairDivideLow(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "HamiltonianFactory" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 44 of file test_hamiltonian_factory.cpp.
References WaveFunctionFactory::buildEmptyTWFForTesting(), OHMMS::Controller, createElectronParticleSet(), doc, HamiltonianFactory::getH(), QMCHamiltonian::getOperatorType(), Libxml2Document::getRoot(), okay, Libxml2Document::parseFromString(), HamiltonianFactory::put(), REQUIRE(), QMCHamiltonian::size(), and QMCHamiltonian::total_size().
| qmcplusplus::TEST_CASE | ( | "convertUPtrToRefvector" | , |
| "" | [type_traits] | ||
| ) |
Definition at line 44 of file test_template_types.cpp.
References CHECK(), convertUPtrToRefVector(), qmcplusplus::Units::charge::e, and qmcplusplus::Units::time::s.
| qmcplusplus::TEST_CASE | ( | "Walker control assign walkers" | , |
| "" | [drivers][walker_control] | ||
| ) |
Definition at line 44 of file test_walker_control.cpp.
References WalkerControlMPI::determineNewWalkerPopulation(), qmcplusplus::Units::mass::me, and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "prime number set 64 bit" | , |
| "" | [utilities] | ||
| ) |
Definition at line 45 of file test_prime_set.cpp.
References PrimeNumberSet< UIntType >::get(), REQUIRE(), and PrimeNumberSet< UIntType >::size().
| qmcplusplus::TEST_CASE | ( | "MultiQuinticSpline" | , |
| "" | [wavefunction][LCAO] | ||
| ) |
Definition at line 46 of file test_multiquintic_spline.cpp.
References MultiQuinticSpline1D< T >::add_spline(), CHECK(), MultiQuinticSpline1D< T >::evaluate(), MultiQuinticSpline1D< T >::initialize(), and OneDimQuinticSpline< Td, Tg, CTd, CTg >::set().
| qmcplusplus::TEST_CASE | ( | "Pade2 functor" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 46 of file test_pade_jastrow.cpp.
References qmcplusplus::Units::distance::A, Pade2ndOrderFunctor< T >::A, B(), Pade2ndOrderFunctor< T >::B, qmcplusplus::Units::charge::C, Pade2ndOrderFunctor< T >::C, CHECK(), Pade2ndOrderFunctor< T >::evaluate(), and Pade2ndOrderFunctor< T >::reset().
| qmcplusplus::TEST_CASE | ( | "ReadFileBuffer_no_file" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 46 of file test_ecp.cpp.
References ReadFileBuffer::open_file(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "gaussian random array length 2" | , |
| "" | [particle_base] | ||
| ) |
Definition at line 46 of file test_random_seq.cpp.
References assignGaussRand(), CHECK(), and qmcplusplus::Units::charge::e.
| qmcplusplus::TEST_CASE | ( | "getSplineBound double" | , |
| "" | [numerics] | ||
| ) |
Definition at line 47 of file test_SplineBound.cpp.
| qmcplusplus::TEST_CASE | ( | "walker assumptions" | , |
| "" | [particle] | ||
| ) |
Currently significant amounts of code assumes that the Walker by default has "Properties" ending with the LOCALPOTENTIAL element.
This opens the door to off by 1 when considering the default size.
Definition at line 47 of file test_walker.cpp.
References ConstantSizeMatrix< T, ALLOC >::cols(), Walker< t_traits, p_traits >::Properties, and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "getSplineBound float" | , |
| "" | [numerics] | ||
| ) |
Definition at line 48 of file test_SplineBound.cpp.
| qmcplusplus::TEST_CASE | ( | "RandomNumberControl random in xml" | , |
| "" | [ohmmsapp] | ||
| ) |
Definition at line 49 of file test_rng_control.cpp.
References OHMMS::Controller, doc, Libxml2Document::getXPathContext(), RandomNumberControl::initialize(), okay, Libxml2Document::parseFromString(), RandomNumberControl::read(), REQUIRE(), and RandomNumberControl::write().
| qmcplusplus::TEST_CASE | ( | "LocalEnergy" | , |
| "" | [estimators] | ||
| ) |
Definition at line 49 of file test_local_energy_est.cpp.
References LocalEnergyEstimator::accumulate(), WalkerConfigurations::begin(), CHECK(), LocalEnergyEstimator::clone(), ParticleSet::create(), MCWalkerConfiguration::createWalkers(), WalkerConfigurations::end(), LocalEnergyEstimator::getName(), REQUIRE(), ScalarEstimatorBase::scalars, and ParticleSet::setName().
| qmcplusplus::TEST_CASE | ( | "BSpline functor one" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 50 of file test_J1_bspline.cpp.
References BsplineFunctor< REAL >::evaluate(), REQUIRE(), and BsplineFunctor< REAL >::resize().
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A Ewald Quasi2D square" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 50 of file test_ewald_quasi2d.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAA::evaluate(), ParticleSet::getSpeciesSet(), lattice, LRCoulombSingleton::QUASI2D, ParticleSet::R, ParticleSet::setName(), LRCoulombSingleton::this_lr_type, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "parsewords" | , |
| "" | [utilities] | ||
| ) |
Definition at line 50 of file test_parser.cpp.
References parsewords(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "test_timer_stack" | , |
| "" | [utilities] | ||
| ) |
Definition at line 51 of file test_timer.cpp.
References TimerManager< TIMER >::createTimer(), TimerManager< TIMER >::current_timer(), REQUIRE(), TimerType< CLOCK >::start(), TimerType< CLOCK >::stop(), and timer_level_coarse.
| qmcplusplus::TEST_CASE | ( | "PerParticleHamiltonianLogger_sum" | , |
| "" | [estimators] | ||
| ) |
Definition at line 51 of file test_PerParticleHamiltonianLogger.cpp.
References CHECK(), PerParticleHamiltonianLogger::collect(), convertUPtrToRefVector(), doc, PerParticleHamiltonianLogger::get_block(), PerParticleHamiltonianLoggerInput::get_to_stdout(), Libxml2Document::getRoot(), node, okay, Libxml2Document::parseFromString(), pset, rank, REQUIRE(), PerParticleHamiltonianLogger::spawnCrowdClone(), PerParticleHamiltonianLogger::startBlock(), walker, and qmcplusplus::hdf::walkers.
| qmcplusplus::TEST_CASE | ( | "FS evaluate" | , |
| "" | [tools] | ||
| ) |
reference numbers and simple_Sk.dat created by fs_ref.py
Definition at line 51 of file test_qmcfstool.cpp.
References CHECK(), and QMCAppBase::parse().
| qmcplusplus::TEST_CASE | ( | "ProjectData::put no series" | , |
| "" | [ohmmsapp] | ||
| ) |
Definition at line 51 of file test_project_data.cpp.
References doc, Libxml2Document::getRoot(), ProjectData::getSeriesIndex(), ProjectData::getTitle(), okay, Libxml2Document::parseFromString(), ProjectData::put(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "OMPclass member" | , |
| "" | [OMP] | ||
| ) |
Definition at line 51 of file test_class_member.cpp.
References maptest< T >::run().
| qmcplusplus::TEST_CASE | ( | "particle_attrib_ops_double" | , |
| "" | [particle_base] | ||
| ) |
Definition at line 51 of file test_attrib_ops.cpp.
| qmcplusplus::TEST_CASE | ( | "QMCUpdate" | , |
| "" | [drivers] | ||
| ) |
Definition at line 51 of file test_clone_manager.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), OHMMS::Controller, ParticleSet::create(), MCWalkerConfiguration::createWalkers(), ParticleSet::getSpeciesSet(), QMCUpdateBase::put(), and ParticleSet::setName().
| qmcplusplus::TEST_CASE | ( | "ParallelExecutor<OPENMP> nested case" | , |
| "" | [concurrency] | ||
| ) |
Definition at line 51 of file test_ParallelExecutorOPENMP.cpp.
References TestTaskOMP().
| qmcplusplus::TEST_CASE | ( | "Spherical Harmonics Many" | , |
| "" | [numerics] | ||
| ) |
Definition at line 52 of file test_ylm.cpp.
References CHECK(), qmcplusplus::Units::distance::m, qmcplusplus::Units::force::N, and Ylm().
| qmcplusplus::TEST_CASE | ( | "array NestedContainers" | , |
| "" | [OhmmsPETE] | ||
| ) |
Definition at line 52 of file test_Array.cpp.
References CHECK(), REQUIRE(), Array< T, D, ALLOC >::resize(), and Array< T, D, ALLOC >::size().
| qmcplusplus::TEST_CASE | ( | "particle_attrib_vector" | , |
| "" | [particle_base] | ||
| ) |
Definition at line 52 of file test_particle_attrib.cpp.
References REQUIRE(), Vector< T, Alloc >::resize(), and Vector< T, Alloc >::size().
| qmcplusplus::TEST_CASE | ( | "OneBodyDensityMatricesInput::copy_construction" | , |
| "" | [estimators] | ||
| ) |
Definition at line 53 of file test_OneBodyDensityMatricesInput.cpp.
References doc, Libxml2Document::getRoot(), node, obdmi, okay, and Libxml2Document::parseFromString().
| qmcplusplus::TEST_CASE | ( | "ReadFileBuffer_simple_serial" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 53 of file test_ecp.cpp.
References ReadFileBuffer::contents(), ReadFileBuffer::length, ReadFileBuffer::open_file(), ReadFileBuffer::read_contents(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "updateXmlNodes" | , |
| "" | [drivers] | ||
| ) |
Definition at line 54 of file test_QMCCostFunctionBase.cpp.
References QMCCostFunctionTest::callUpdateXmlNodes(), comm, OHMMS::Controller, doc, QMCCostFunctionTest::getDoc(), Libxml2Document::getRoot(), getXMLAttributeValue(), okay, Libxml2Document::parseFromString(), REQUIRE(), QMCCostFunctionBase::setRootName(), and QMCCostFunctionBase::setWaveFunctionNode().
| qmcplusplus::TEST_CASE | ( | "accumulator basic float" | , |
| "" | [estimators] | ||
| ) |
Definition at line 54 of file test_accumulator.cpp.
| qmcplusplus::TEST_CASE | ( | "test_communicate_split_two" | , |
| "" | [message] | ||
| ) |
Definition at line 54 of file test_communciate.cpp.
References OHMMS::Controller, REQUIRE(), and Communicate::size().
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A Ewald2D body center" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 54 of file test_ewald2d.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), dot(), CoulombPBCAA::evaluate(), ParticleSet::getSpeciesSet(), lattice, ParticleSet::R, ParticleSet::setName(), LRCoulombSingleton::STRICT2D, LRCoulombSingleton::this_lr_type, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "walker HDF read and write" | , |
| "" | [particle] | ||
| ) |
Definition at line 54 of file test_walker.cpp.
References Communicate::barrier(), OHMMS::Controller, WalkerConfigurations::createWalkers(), HDFWalkerOutput::dump(), WalkerConfigurations::getActiveWalkers(), okay, Walker< t_traits, p_traits >::R, HDFWalkerInput_0_4::read_hdf5(), REQUIRE(), Communicate::setName(), WalkerConfigurations::setWalkerOffsets(), Communicate::size(), and qmcplusplus::hdf::version.
| qmcplusplus::TEST_CASE | ( | "ReferencePointsInput::makeReferencePointsInput" | , |
| "" | [estimators] | ||
| ) |
Definition at line 54 of file test_ReferencePointsInput.cpp.
References CHECK(), doc, Libxml2Document::getRoot(), makeReferencePointsInput(), node, okay, and Libxml2Document::parseFromString().
| qmcplusplus::TEST_CASE | ( | "FakeRandom noops" | , |
| "" | [utilities] | ||
| ) |
Definition at line 54 of file test_FakeRandom.cpp.
References CHECK(), and FakeRandom< T >::set_value().
| qmcplusplus::TEST_CASE | ( | "spline_function_3" | , |
| "" | [numerics] | ||
| ) |
Definition at line 55 of file test_one_dim_cubic_spline.cpp.
References CHECK(), CubicSplineSolve(), and n.
| qmcplusplus::TEST_CASE | ( | "solveGeneralizedEigenvaluesInv" | , |
| "" | [drivers] | ||
| ) |
Definition at line 55 of file test_Eigensolver.cpp.
References app_log(), CHECK(), qmcplusplus::Units::force::N, and Eigensolver::solveGeneralizedEigenvalues_Inv().
| qmcplusplus::TEST_CASE | ( | "TraceManager check_trace_build" | , |
| "" | [estimators] | ||
| ) |
Definition at line 55 of file test_trace_manager.cpp.
References TraceSamples< T >::checkout_array(), TraceManager::checkout_complex(), TraceManager::checkout_int(), TraceManager::checkout_real(), and ParticleSet::create().
| qmcplusplus::TEST_CASE | ( | "Density Estimator evaluate exception" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 56 of file test_density_estimator.cpp.
References DensityEstimator::evaluate(), DensityEstimator::getClassName(), REQUIRE(), and DensityEstimator::resetTargetParticleSet().
| qmcplusplus::TEST_CASE | ( | "accumulator basic double" | , |
| "" | [estimators] | ||
| ) |
Definition at line 56 of file test_accumulator.cpp.
| qmcplusplus::TEST_CASE | ( | "Vector simple intializer list" | , |
| "" | [OhmmsPETE] | ||
| ) |
Definition at line 56 of file test_Vector.cpp.
References CHECK().
| qmcplusplus::TEST_CASE | ( | "SpaceGridInputs::parseXML::axes" | , |
| "" | [estimators] | ||
| ) |
Definition at line 57 of file test_SpaceGridInput.cpp.
References CHECK(), checkVec(), doc, SpaceGridInput::get_axis_grids(), SpaceGridInput::get_axis_labels(), SpaceGridInput::get_axis_p1s(), SpaceGridInput::get_axis_scales(), SpaceGridInput::get_origin_fraction(), SpaceGridInput::get_origin_p1(), SpaceGridInput::get_origin_p2(), Libxml2Document::getRoot(), node, okay, Libxml2Document::parseFromString(), REQUIRE(), and ValidSpinDensityInput::xml.
| qmcplusplus::TEST_CASE | ( | "kcontainer at twist in 3D" | , |
| "" | [longrange] | ||
| ) |
Definition at line 57 of file test_kcontainer.cpp.
References CHECK(), qmcplusplus::Units::charge::e, KContainer::kpts, KContainer::kpts_cart, lattice, and KContainer::updateKLists().
| qmcplusplus::TEST_CASE | ( | "VectorSoaContainer copy constructor" | , |
| "" | [OhmmsSoA] | ||
| ) |
Definition at line 57 of file test_vector_soa.cpp.
References CHECK(), and VectorSoaContainer< T, D, Alloc >::copyIn().
| qmcplusplus::TEST_CASE | ( | "StdRandom clone" | , |
| "" | [utilities] | ||
| ) |
Definition at line 57 of file test_StdRandom.cpp.
References CHECK(), and StdRandom< T >::init().
| qmcplusplus::TEST_CASE | ( | "EstimatorManagerCrowd PerParticleHamiltonianLogger integration" | , |
| "" | [estimators] | ||
| ) |
Definition at line 58 of file test_EstimatorManagerCrowd.cpp.
References EstimatorManagerCrowd::accumulate(), EstimatorManagerInput::append(), ProjectData::BATCH, CHECK(), comm, OHMMS::Controller, convertUPtrToRefVector(), qmcplusplus::testing::createEstimatorManagerNewVMCInputXML(), emi(), emn, estimators_doc, RefVectorWithLeader< T >::getLeader(), EstimatorManagerNew::getNumEstimators(), EstimatorManagerNew::getNumScalarEstimators(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), ham, hamiltonian_pool, QMCHamiltonian::informOperatorsOfListener(), MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), MinimalHamiltonianPool::make_hamWithEE(), QMCHamiltonian::mw_evaluate(), ParticleSet::mw_update(), qmcplusplus::hdf::num_walkers, particle_pool, pset, EstimatorManagerCrowd::registerListeners(), QMCHamiltonian::saveProperty(), test_project, twf, walker, qmcplusplus::hdf::walkers, and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "NonLocalECPotential" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 58 of file test_NonLocalECPotential.cpp.
References SpeciesSet::addAttribute(), NonLocalECPotential::addComponent(), SpeciesSet::addSpecies(), ParticleSet::addTable(), Matrix< T, Alloc >::begin(), CHECK(), Matrix< T, Alloc >::cols(), comm, OHMMS::Controller, TestNonLocalECPotential::copyGridUnrotatedForTest(), ParticleSet::create(), NonLocalECPotential::createResource(), ParticleSet::createResource(), ParticleSet::createSK(), TestNonLocalECPotential::didGridChange(), qmcplusplus::testing::getParticularListener(), ParticleSet::getSpeciesSet(), lattice, NonLocalECPotential::makeClone(), TestNonLocalECPotential::mw_evaluateImpl(), qmcplusplus::hdf::num_walkers, okay, ECPComponentBuilder::pp_nonloc, ParticleSet::R, ECPComponentBuilder::read_pp_file(), REQUIRE(), ParticleSet::resetGroups(), ParticleSet::setName(), NonLocalECPotential::setRandomGenerator(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "DiracMatrix_inverse" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 58 of file test_DiracMatrix.cpp.
References CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), Matrix< T, Alloc >::cols(), Matrix< T, Alloc >::data(), TestMatrix1::fillInput(), TestMatrix1::fillInverse(), DiracMatrix< T_FP >::invert_transpose(), TestMatrix1::logDet(), Matrix< T, Alloc >::resize(), Matrix< T, Alloc >::rows(), and qmcplusplus::simd::transpose().
| qmcplusplus::TEST_CASE | ( | "gaussian random array length 3" | , |
| "" | [particle_base] | ||
| ) |
Definition at line 58 of file test_random_seq.cpp.
References assignGaussRand(), CHECK(), and qmcplusplus::Units::charge::e.
| qmcplusplus::TEST_CASE | ( | "FairDivideLow_three" | , |
| "" | [utilities] | ||
| ) |
Definition at line 58 of file test_partition.cpp.
References FairDivideLow(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "Estimator adhoc addVector operator" | , |
| "" | [estimators] | ||
| ) |
Definition at line 60 of file test_manager.cpp.
References REQUIRE().
| qmcplusplus::TEST_CASE | ( | "periodic_bulk_bconds" | , |
| "" | [lattice] | ||
| ) |
Lattice is defined but Open BC is also used.
Definition at line 60 of file test_ParticleBConds.cpp.
References DTD_BConds< T, D, SC >::apply_bc(), CrystalLattice< T, D >::BoxBConds, CHECK(), Tensor< T, D >::diagonal(), OHMMS_PRECISION, CrystalLattice< T, D >::R, REQUIRE(), CrystalLattice< T, D >::reset(), and CrystalLattice< T, D >::Volume.
| qmcplusplus::TEST_CASE | ( | "MomentumDistribution::MomentumDistribution" | , |
| "" | [estimators] | ||
| ) |
Definition at line 61 of file test_MomentumDistribution.cpp.
References ProjectData::BATCH, CHECK(), comm, OHMMS::Controller, crowd, doc, Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), lattice, MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), qmcplusplus::testing::makeTestLattice(), node, oeba, okay, Libxml2Document::parseFromString(), particle_pool, pset, test_project, MomentumDistributionTests::testCopyConstructor(), and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "Gaussian Combo" | , |
| "" | [numerics] | ||
| ) |
Definition at line 61 of file test_gaussian_basis.cpp.
References GaussianCombo< T >::addGaussian(), CHECK(), GaussianCombo< T >::d2Y, GaussianCombo< T >::d3Y, GaussianCombo< T >::df(), GaussianCombo< T >::dY, GaussianCombo< T >::evaluate(), GaussianCombo< T >::evaluateAll(), GaussianCombo< T >::evaluateWithThirdDeriv(), GaussianCombo< T >::f(), REQUIRE(), GaussianCombo< T >::size(), and GaussianCombo< T >::Y.
| qmcplusplus::TEST_CASE | ( | "test_loop_control" | , |
| "" | [utilities] | ||
| ) |
Definition at line 62 of file test_runtime_manager.cpp.
References RunTimeControl< CLOCK >::checkStop(), RunTimeControl< CLOCK >::generateStopMessage(), REQUIRE(), RunTimeControl< CLOCK >::runtime_padding(), LoopTimer< CLOCK >::start(), and LoopTimer< CLOCK >::stop().
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A BCC H Ewald3D" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 62 of file test_coulomb_pbcAA_ewald.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAA::evalConsts(), CoulombPBCAA::evaluate(), ParticleSet::getSpeciesSet(), lattice, ParticleSet::R, and ParticleSet::setName().
| qmcplusplus::TEST_CASE | ( | "integrateListeners" | , |
| "" | [hamiltonian] | ||
| ) |
QMCHamiltonian + Hamiltonians with listeners integration test.
Definition at line 62 of file test_QMCHamiltonian.cpp.
References Matrix< T, Alloc >::begin(), CHECK(), comm, OHMMS::Controller, convertUPtrToRefVector(), copy(), Matrix< T, Alloc >::end(), generate_test_data, qmcplusplus::testing::getParticularListener(), qmcplusplus::testing::getSummingListener(), hamiltonian_pool, MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), MinimalHamiltonianPool::makeHamWithEEEI(), QMCHamiltonian::mw_evaluate(), QMCHamiltonian::mw_registerKineticListener(), QMCHamiltonian::mw_registerLocalEnergyListener(), QMCHamiltonian::mw_registerLocalIonPotentialListener(), QMCHamiltonian::mw_registerLocalPotentialListener(), ParticleSet::mw_update(), qmcplusplus::hdf::num_walkers, particle_pool, pset_target, and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "VMCBatched::calc_default_local_walkers" | , |
| "" | [drivers] | ||
| ) |
Definition at line 62 of file test_VMCBatched.cpp.
References ProjectData::BATCH, test_project, and VMCBatchedTest::testCalcDefaultLocalWalkers().
| qmcplusplus::TEST_CASE | ( | "Pade Jastrow" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 63 of file test_pade_jastrow.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), RadialJastrowBuilder::buildComponent(), CHECK(), OHMMS::Controller, ParticleSet::create(), doc, ParticleSet::G, Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), ParticleSet::L, okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "DiracMatrixComputeCUDA_different_batch_sizes" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 63 of file test_DiracMatrixComputeCUDA.cpp.
References qmcplusplus::Units::distance::A, CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), qmcplusplus::syclBLAS::copy_n(), Matrix< T, Alloc >::data(), DiracMatrixComputeCUDA< VALUE_FP >::invert_transpose(), log_values(), DiracMatrixComputeCUDA< VALUE_FP >::mw_invertTranspose(), queue, and Matrix< T, Alloc >::resize().
| qmcplusplus::TEST_CASE | ( | "OutputManager output" | , |
| "" | [utilities] | ||
| ) |
Definition at line 63 of file test_output_manager.cpp.
References app_debug, app_error(), app_log(), app_out, app_summary(), DEBUG, debug_out, err_out, HIGH, init_string_output(), LOW, outputManager, REQUIRE(), reset_string_output(), OutputManagerClass::setVerbosity(), and summary_out.
| qmcplusplus::TEST_CASE | ( | "symmetric_distance_table OpenBC" | , |
| "" | [particle] | ||
| ) |
make sure getCoordinates().getAllParticlePos() is updated no matter SoA or AoS.
Definition at line 64 of file test_ParticleSet.cpp.
References ParticleSet::addTable(), CHECK(), ParticleSet::create(), DynamicCoordinates::getAllParticlePos(), ParticleSet::getCoordinates(), DistanceTableAA::getDistances(), ParticleSet::getDistTableAA(), ParticleSet::R, ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "applyCuspInfo" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 65 of file test_soa_cusp_corr.cpp.
References LCAOrbitalSet::C, CHECK(), computeRadialPhiBar(), OHMMS::Controller, SPOSetBuilderFactory::createSPOSetBuilder(), doc, SPOSet::getOrbitalSetSize(), Libxml2Document::getRoot(), Libxml2Document::getXPathContext(), LCAOrbitalSet::isIdentity(), LCAOrbitalSet::isOMPoffload(), LCAOrbitalSet::myBasisSet, okay, Libxml2Document::parse(), readCuspInfo(), XMLParticleParser::readXML(), removeSTypeOrbitals(), REQUIRE(), Matrix< T, Alloc >::resize(), Vector< T, Alloc >::resize(), and splitPhiEta().
| qmcplusplus::TEST_CASE | ( | "Vector nested intializer list" | , |
| "" | [OhmmsPETE] | ||
| ) |
Definition at line 65 of file test_Vector.cpp.
References CHECK(), and qmcplusplus::Units::second.
| qmcplusplus::TEST_CASE | ( | "EstimatorManagerInput::readXML" | , |
| "" | [estimators] | ||
| ) |
Definition at line 66 of file test_EstimatorManagerInput.cpp.
References Libxml2Document::addChild(), CHECK(), qmcplusplus::testing::createEstimatorManagerNewInputXML(), doc, emi(), estimators_doc, EstimatorManagerInput::get_estimator_inputs(), EstimatorManagerInput::get_scalar_estimator_inputs(), Libxml2Document::getRoot(), node, okay, Libxml2Document::parseFromString(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "test_timer_scoped" | , |
| "" | [utilities] | ||
| ) |
Definition at line 67 of file test_timer.cpp.
References CHECK(), TimerManager< TIMER >::createTimer(), TimerType< CLOCK >::get_num_calls(), TimerType< CLOCK >::get_total(), REQUIRE(), and timer_level_coarse.
| qmcplusplus::TEST_CASE | ( | "Pair Correlation" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 67 of file test_PairCorrEstimator.cpp.
References PairCorrEstimator::addObservables(), ParticleSet::addTable(), ParticleSet::Collectables, OHMMS::Controller, doc, PairCorrEstimator::evaluate(), ParticleSet::get(), ParticleSet::getDistTable(), ParticleSet::getLattice(), OhmmsElementBase::getName(), ParticleSetPool::getParticleSet(), Libxml2Document::getRoot(), gofr_xml, ParticleSet::isSameMass(), lat_xml, Libxml2Document::parseFromString(), ParticleSet::PropertyList, pset_xml, ParticleSetPool::put(), PairCorrEstimator::put(), ParticleSet::R, ParticleSetPool::readSimulationCellXML(), REQUIRE(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "ProjectData::put with series" | , |
| "" | [ohmmsapp] | ||
| ) |
Definition at line 67 of file test_project_data.cpp.
References doc, Libxml2Document::getRoot(), ProjectData::getSeriesIndex(), ProjectData::getTitle(), okay, Libxml2Document::parseFromString(), ProjectData::put(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "ConstantSizeMatrix Ohmms integration" | , |
| "" | [containers] | ||
| ) |
Definition at line 67 of file test_ConstantSizeMatrix.cpp.
References CHECK(), and ConstantSizeMatrix< T, ALLOC >::size().
| qmcplusplus::TEST_CASE | ( | "cuBLAS_LU::computeLogDet" | , |
| "" | [wavefunction][CUDA] | ||
| ) |
Single double computeLogDet.
Definition at line 67 of file test_cuBLAS_LU.cpp.
References batch_size, CHECK(), qmcplusplus::cuBLAS_LU::computeLogDet_batched(), cudaCheck, cudaMemcpyAsync, cudaMemcpyDeviceToHost, cudaMemcpyHostToDevice, cudaStreamSynchronize, dev_log_values(), dev_lu(), dev_lus(), dev_pivots(), hstream, lda, log_values(), lu, lus, n, and pivots.
| qmcplusplus::TEST_CASE | ( | "DiracMatrixComputeCUDA_large_determinants_benchmark_legacy_1024_4" | , |
| "" | [wavefunction][fermion][.benchmark] | ||
| ) |
This and other [.benchmark] benchmarks only run if "[benchmark]" is explicitly passed as tag to test.
Definition at line 68 of file benchmark_DiracMatrixComputeCUDA.cpp.
References DiracComputeBenchmarkParameters::batch_size, DiracMatrix< T_FP >::invert_transpose(), log_values(), qmcplusplus::testing::makeRngSpdMatrix(), qmcplusplus::Units::meter, DiracMatrixComputeCUDA< VALUE_FP >::mw_invertTranspose(), DiracComputeBenchmarkParameters::n, DiracComputeBenchmarkParameters::name, queue, and DiracComputeBenchmarkParameters::str().
| qmcplusplus::TEST_CASE | ( | "ReadFileBuffer_simple_mpi" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 68 of file test_ecp.cpp.
References ReadFileBuffer::contents(), OHMMS::Controller, ReadFileBuffer::length, ReadFileBuffer::open_file(), ReadFileBuffer::read_contents(), and REQUIRE().
Definition at line 68 of file test_QMCDriverFactory.cpp.
References CHECK(), comm, OHMMS::Controller, qmcplusplus::testing::createDriver(), doc, Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), QMCDriverFactory::DriverAssemblyState::new_run_type, node, okay, Libxml2Document::parseFromString(), QMCDriverFactory::readSection(), REQUIRE(), test_project, qmcplusplus::testing::valid_vmc_input_sections, qmcplusplus::testing::valid_vmc_input_vmc_index, and VMC.
| qmcplusplus::TEST_CASE | ( | "Test OptimizableObject" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 69 of file test_OptimizableObject.cpp.
References CHECK(), and UniqueOptObjRefs::push_back().
| qmcplusplus::TEST_CASE | ( | "gaussian random particle attrib array length 1" | , |
| "" | [particle_base] | ||
| ) |
Definition at line 71 of file test_random_seq.cpp.
References CHECK(), makeGaussRandomWithEngine(), and Vector< T, Alloc >::resize().
| qmcplusplus::TEST_CASE | ( | "spline_function_4" | , |
| "" | [numerics] | ||
| ) |
Definition at line 71 of file test_one_dim_cubic_spline.cpp.
References CHECK(), CubicSplineSolve(), qmcplusplus::Units::charge::e, and n.
| qmcplusplus::TEST_CASE | ( | "ResourceCollection" | , |
| "" | [utilities] | ||
| ) |
Definition at line 72 of file test_ResourceCollection.cpp.
References CHECK(), WFCResourceConsumer::createResource(), MemoryResource::data, WFCResourceConsumer::getResourceHandle(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "MCPopulation::createWalkers_walker_ids" | , |
| "" | [particle][population] | ||
| ) |
Definition at line 73 of file test_MCPopulation.cpp.
References CHECK(), comm, OHMMS::Controller, hamiltonian_pool, MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), MinimalHamiltonianPool::make_hamWithEE(), num_ranks, particle_pool, twf, and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "ewald3d df" | , |
| "" | [lrhandler] | ||
| ) |
evalaute bare Coulomb derivative using EwaldHandler3D
Definition at line 74 of file test_ewald3d.cpp.
References CrystalLattice< T, D >::BoxBConds, CHECK(), ParticleSet::createSK(), Tensor< T, D >::diagonal(), EwaldHandler3D::evaluate(), EwaldHandler3D::evaluateLR(), EwaldHandler3D::initBreakup(), LRHandlerBase::LR_kc, LRHandlerBase::LR_rc, EwaldHandler3D::lrDf(), LRHandlerBase::MaxKshell, CrystalLattice< T, D >::R, CrystalLattice< T, D >::reset(), CrystalLattice< T, D >::Rv, EwaldHandler3D::Sigma, sqrt(), EwaldHandler3D::srDf(), and CrystalLattice< T, D >::Volume.
| qmcplusplus::TEST_CASE | ( | "tiny vector" | , |
| "" | [OhmmsPETE] | ||
| ) |
Definition at line 75 of file test_TinyVector.cpp.
| qmcplusplus::TEST_CASE | ( | "VectorViewer" | , |
| "" | [OhmmsPETE] | ||
| ) |
Definition at line 75 of file test_Vector.cpp.
References REQUIRE(), and Vector< T, Alloc >::size().
| qmcplusplus::TEST_CASE | ( | "Array::data" | , |
| "" | [OhmmsPETE] | ||
| ) |
Definition at line 76 of file test_Array.cpp.
References CHECK(), Array< T, D, ALLOC >::data(), Array< T, D, ALLOC >::data_at(), REQUIRE(), and Array< T, D, ALLOC >::size().
| qmcplusplus::TEST_CASE | ( | "bracket minimum" | , |
| "" | [numerics] | ||
| ) |
Definition at line 77 of file test_min_oned.cpp.
References MinTest::find_bracket(), and MinTest::find_bracket_bound().
| qmcplusplus::TEST_CASE | ( | "SmallMatrixDetCalculator::evaluate-Small" | , |
| "" | [wavefunction][fermion][multidet] | ||
| ) |
Simple synthetic test case will trip on changes in this method.
Definition at line 78 of file test_multi_dirac_determinant.cpp.
References CHECK(), TestSmallMatrixDetCalculator< T >::customized_evaluate(), and TestSmallMatrixDetCalculator< T >::generic_evaluate().
| qmcplusplus::TEST_CASE | ( | "FakeRandom clone" | , |
| "" | [utilities] | ||
| ) |
Definition at line 78 of file test_FakeRandom.cpp.
References CHECK(), and FakeRandom< T >::write().
| qmcplusplus::TEST_CASE | ( | "Crowd integration" | , |
| "" | [drivers] | ||
| ) |
Definition at line 79 of file test_Crowd.cpp.
References comm, OHMMS::Controller, crowd, SetupPools::hamiltonian_pool, SetupPools::particle_pool, pools, and SetupPools::wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A BCC H" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 79 of file test_coulomb_pbcAA.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAA::evalConsts(), CoulombPBCAA::evaluate(), CoulombPBCAA::get_madelung_constant(), ParticleSet::getSpeciesSet(), lattice, ParticleSet::R, and ParticleSet::setName().
| qmcplusplus::TEST_CASE | ( | "VectorSoaContainer move constructor" | , |
| "" | [OhmmsSoA] | ||
| ) |
Definition at line 79 of file test_vector_soa.cpp.
References CHECK(), and VectorSoaContainer< T, D, Alloc >::copyIn().
| qmcplusplus::TEST_CASE | ( | "ParticleSetPool random" | , |
| "" | [qmcapp] | ||
| ) |
Definition at line 79 of file test_particle_pool.cpp.
References OHMMS::Controller, doc, ParticleSetPool::getParticleSet(), Libxml2Document::getRoot(), ParticleSet::isSpinor(), okay, Libxml2Document::parseFromString(), ParticleSetPool::put(), ParticleSetPool::randomize(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "FairDivideLow_four" | , |
| "" | [utilities] | ||
| ) |
Definition at line 80 of file test_partition.cpp.
References FairDivideLow(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "LocalEnergy with hdf5" | , |
| "" | [estimators] | ||
| ) |
Definition at line 80 of file test_local_energy_est.cpp.
References LocalEnergyEstimator::add2Record(), WalkerConfigurations::begin(), hdf_archive::close(), hdf_archive::create(), ParticleSet::create(), MCWalkerConfiguration::createWalkers(), LocalEnergyEstimator::registerObservables(), REQUIRE(), and ParticleSet::setName().
| qmcplusplus::TEST_CASE | ( | "srcoul df" | , |
| "" | [lrhandler] | ||
| ) |
evalaute bare Coulomb derivative in 3D
Definition at line 81 of file test_srcoul.cpp.
References CrystalLattice< T, D >::BoxBConds, CHECK(), ParticleSet::createSK(), EslerCoulomb3D_ForSRCOUL::df(), Tensor< T, D >::diagonal(), LRHandlerSRCoulomb< Func, BreakupBasis >::evaluate(), LRHandlerSRCoulomb< Func, BreakupBasis >::evaluateLR(), LRHandlerSRCoulomb< Func, BreakupBasis >::initBreakup(), LRHandlerBase::LR_kc, LRHandlerBase::LR_rc, LRHandlerSRCoulomb< Func, BreakupBasis >::lrDf(), LRHandlerBase::MaxKshell, CrystalLattice< T, D >::R, EslerCoulomb3D_ForSRCOUL::reset(), CrystalLattice< T, D >::reset(), CrystalLattice< T, D >::Rv, LRHandlerSRCoulomb< Func, BreakupBasis >::srDf(), and CrystalLattice< T, D >::Volume.
| qmcplusplus::TEST_CASE | ( | "matrix converting assignment" | , |
| "" | [OhmmsPETE] | ||
| ) |
Definition at line 81 of file test_Matrix.cpp.
References Matrix< T, Alloc >::assignUpperLeft(), and CHECK().
| qmcplusplus::TEST_CASE | ( | "CloneManager" | , |
| "" | [drivers] | ||
| ) |
Definition at line 81 of file test_clone_manager.cpp.
References OHMMS::Controller.
| qmcplusplus::TEST_CASE | ( | "uniform 3D Lattice layout" | , |
| "" | [lattice] | ||
| ) |
Definition at line 81 of file test_ParticleBConds.cpp.
References CrystalLattice< T, D >::BoxBConds, Tensor< T, D >::diagonal(), CrystalLattice< T, D >::R, REQUIRE(), and CrystalLattice< T, D >::reset().
| qmcplusplus::TEST_CASE | ( | "ReadFileBuffer_ecp" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 82 of file test_ecp.cpp.
References OHMMS::Controller, okay, ECPComponentBuilder::read_pp_file(), REQUIRE(), and ECPComponentBuilder::Zeff.
| qmcplusplus::TEST_CASE | ( | "TwoBodyJastrow two variables" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 82 of file test_J2_derivatives.cpp.
References TwoBodyJastrow< FT >::addFunc(), CHECK(), TwoBodyJastrow< FT >::checkOutVariables(), TwoBodyJastrow< FT >::extractOptimizableObjectRefs(), get_two_species_particleset(), TwoBodyJastrow< FT >::getComponentOffset(), VariableSet::insertFrom(), REQUIRE(), VariableSet::resetIndex(), and VariableSet::size_of_active().
| qmcplusplus::TEST_CASE | ( | "gaussian random input one" | , |
| "" | [particle_base] | ||
| ) |
Definition at line 82 of file test_random_seq.cpp.
References assignGaussRand(), CHECK(), and FakeRandom< T >::set_value().
| qmcplusplus::TEST_CASE | ( | "ModernStringUtils_string2Real" | , |
| "" | [utilities] | ||
| ) |
Definition at line 83 of file test_ModernStringUtils.cpp.
References CHECK().
| qmcplusplus::TEST_CASE | ( | "WaveFunctionPool" | , |
| "" | [qmcapp] | ||
| ) |
Definition at line 83 of file test_wavefunction_pool.cpp.
References ProjectData::BATCH, OHMMS::Controller, doc, TrialWaveFunction::getOrbitals(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), okay, Libxml2Document::parseFromString(), REQUIRE(), setupParticleSetPool(), and test_project.
| qmcplusplus::TEST_CASE | ( | "DiracMatrix_inverse_matching" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 83 of file test_DiracMatrix.cpp.
References CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), Matrix< T, Alloc >::cols(), Matrix< T, Alloc >::data(), DiracMatrix< T_FP >::invert_transpose(), Matrix< T, Alloc >::resize(), Matrix< T, Alloc >::rows(), and qmcplusplus::simd::transpose().
| qmcplusplus::TEST_CASE | ( | "SPO input spline from xml LiH_msd" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 83 of file test_spo_collection_input_MSD_LCAO_h5.cpp.
References app_log(), and test_LiH_msd_xml_input().
| qmcplusplus::TEST_CASE | ( | "convertPtrToRefvectorSubset" | , |
| "" | [type_traits] | ||
| ) |
Definition at line 84 of file test_template_types.cpp.
References CHECK(), and convertPtrToRefVectorSubset().
| qmcplusplus::TEST_CASE | ( | "accumulator some values float" | , |
| "" | [estimators] | ||
| ) |
Definition at line 85 of file test_accumulator.cpp.
| qmcplusplus::TEST_CASE | ( | "ProjectData::TestDriverVersion" | , |
| "" | [ohmmsapp] | ||
| ) |
Definition at line 85 of file test_project_data.cpp.
References doc, ProjectData::getDriverVersion(), Libxml2Document::getRoot(), ProjectData::getSeriesIndex(), okay, Libxml2Document::parseFromString(), ProjectData::put(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A Ewald Quasi2D triangular" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 85 of file test_ewald_quasi2d.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAA::evaluate(), ParticleSet::getSpeciesSet(), lattice, LRCoulombSingleton::QUASI2D, ParticleSet::R, ParticleSet::setName(), sqrt(), LRCoulombSingleton::this_lr_type, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "tiny vector operator out" | , |
| "" | [OhmmsPETE] | ||
| ) |
Definition at line 86 of file test_TinyVector.cpp.
References CHECK().
| qmcplusplus::TEST_CASE | ( | "particle_attrib_ops_complex" | , |
| "" | [particle_base] | ||
| ) |
Definition at line 86 of file test_attrib_ops.cpp.
| qmcplusplus::TEST_CASE | ( | "OptimizableObject HDF output and input" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 87 of file test_OptimizableObject.cpp.
References CHECK(), FakeOptimizableObject::checkInVariablesExclusive(), FakeOptimizableObject::extra_data, VariableSet::readFromHDF(), FakeOptimizableObject::readVariationalParameters(), VariableSet::writeToHDF(), and FakeOptimizableObject::writeVariationalParameters().
| qmcplusplus::TEST_CASE | ( | "accumulator some values double" | , |
| "" | [estimators] | ||
| ) |
Definition at line 87 of file test_accumulator.cpp.
| qmcplusplus::TEST_CASE | ( | "kcontainer at gamma in 2D" | , |
| "" | [longrange] | ||
| ) |
Definition at line 87 of file test_kcontainer.cpp.
References CHECK(), qmcplusplus::Units::charge::e, KContainer::kpts, KContainer::kpts_cart, lattice, sqrt(), and KContainer::updateKLists().
| qmcplusplus::TEST_CASE | ( | "Array::dimension sizes constructor" | , |
| "" | [OhmmsPETE] | ||
| ) |
Definition at line 87 of file test_Array.cpp.
References CHECK(), and Array< T, D, ALLOC >::shape().
| qmcplusplus::TEST_CASE | ( | "solveGeneralizedEigenvaluesCompare" | , |
| "" | [drivers] | ||
| ) |
Definition at line 87 of file test_Eigensolver.cpp.
References CHECK(), qmcplusplus::Units::force::N, Eigensolver::solveGeneralizedEigenvalues(), and Eigensolver::solveGeneralizedEigenvalues_Inv().
| qmcplusplus::TEST_CASE | ( | "spline_function_5" | , |
| "" | [numerics] | ||
| ) |
Definition at line 88 of file test_one_dim_cubic_spline.cpp.
References CHECK(), CubicSplineSolve(), and n.
| qmcplusplus::TEST_CASE | ( | "WalkerControl round trip index conversions" | , |
| "" | [drivers][walker_control] | ||
| ) |
Definition at line 89 of file test_walker_control.cpp.
References REQUIRE().
| qmcplusplus::TEST_CASE | ( | "Bare KE Pulay PBC" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 89 of file test_bare_kinetic.cpp.
References SpeciesSet::addAttribute(), TrialWaveFunction::addComponent(), SpeciesSet::addSpecies(), RadialJastrowBuilder::buildComponent(), CHECK(), OHMMS::Controller, ParticleSet::create(), ParticleSet::createSK(), doc, BareKineticEnergy::evaluate(), TrialWaveFunction::evaluateLog(), BareKineticEnergy::evaluateWithIonDerivs(), Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), ParticleSet::getTotalNum(), lattice, okay, Libxml2Document::parseFromString(), BareKineticEnergy::put(), ParticleSet::R, REQUIRE(), ParticleSet::resetGroups(), Vector< T, Alloc >::resize(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "HamiltonianFactory pseudopotential" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 89 of file test_hamiltonian_factory.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), WaveFunctionFactory::buildEmptyTWFForTesting(), OHMMS::Controller, createElectronParticleSet(), doc, Libxml2Document::getRoot(), okay, Libxml2Document::parseFromString(), HamiltonianFactory::put(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "FairDivideAligned" | , |
| "" | [utilities] | ||
| ) |
Definition at line 90 of file test_partition.cpp.
References FairDivideAligned(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "updateXmlNodes with existing element" | , |
| "" | [drivers] | ||
| ) |
Definition at line 90 of file test_QMCCostFunctionBase.cpp.
References QMCCostFunctionTest::callUpdateXmlNodes(), comm, OHMMS::Controller, doc, QMCCostFunctionTest::getDoc(), Libxml2Document::getRoot(), getXMLAttributeValue(), okay, Libxml2Document::parseFromString(), REQUIRE(), QMCCostFunctionBase::setRootName(), and QMCCostFunctionBase::setWaveFunctionNode().
| qmcplusplus::TEST_CASE | ( | "ModernStringUtils_string2Int" | , |
| "" | [utilities] | ||
| ) |
Definition at line 90 of file test_ModernStringUtils.cpp.
References CHECK().
| qmcplusplus::TEST_CASE | ( | "test_timer_flat_profile" | , |
| "" | [utilities] | ||
| ) |
Definition at line 91 of file test_timer.cpp.
References TimerManager< TIMER >::FlatProfileData::callList, CHECK(), TimerManager< TIMER >::collate_flat_profile(), TimerManager< TIMER >::createTimer(), TimerManager< TIMER >::FlatProfileData::nameList, REQUIRE(), set_num_calls(), set_total_time(), and TimerManager< TIMER >::FlatProfileData::timeList.
| qmcplusplus::TEST_CASE | ( | "symmetric_distance_table PBC" | , |
| "" | [particle] | ||
| ) |
Definition at line 91 of file test_ParticleSet.cpp.
References ParticleSet::addTable(), CrystalLattice< T, D >::BoxBConds, CHECK(), ParticleSet::create(), DistanceTableAA::getDistances(), ParticleSet::getDistTableAA(), ParticleSet::R, CrystalLattice< T, D >::R, CrystalLattice< T, D >::reset(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "read_particleset_recorder_xml" | , |
| "" | [particle_io][xml] | ||
| ) |
Definition at line 91 of file test_xml_io.cpp.
References CHECK(), doc, Libxml2Document::getRoot(), okay, Libxml2Document::parseFromString(), XMLParticleParser::readXML(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "EstimatorManagerNew::collectMainEstimators" | , |
| "" | [estimators] | ||
| ) |
Definition at line 92 of file test_EstimatorManagerNew.cpp.
References CHECK(), EstimatorManagerNewTest::collectMainEstimators(), OHMMS::Controller, EstimatorManagerNewTest::em, embt, EstimatorManagerNewTest::fakeMainScalarSamples(), EstimatorManagerNew::get_AverageCache(), ham, REQUIRE(), and EstimatorManagerNewTest::testReplaceMainEstimator().
| qmcplusplus::TEST_CASE | ( | "QMCDriverFactory create VMCBatched driver" | , |
| "" | [qmcapp] | ||
| ) |
Definition at line 92 of file test_QMCDriverFactory.cpp.
References ProjectData::BATCH, CHECK(), comm, OHMMS::Controller, qmcplusplus::testing::createDriver(), doc, Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), QMCDriverFactory::DriverAssemblyState::new_run_type, node, okay, Libxml2Document::parseFromString(), QMCDriverFactory::readSection(), REQUIRE(), test_project, qmcplusplus::testing::valid_vmc_batch_input_vmc_batch_index, qmcplusplus::testing::valid_vmc_input_sections, qmcplusplus::testing::valid_vmc_input_vmc_batch_index, and VMC_BATCH.
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-B BCC H" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 94 of file test_coulomb_pbcAB.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAB::evalConsts(), CoulombPBCAA::evaluate(), CoulombPBCAB::evaluate(), ParticleSet::getSpeciesSet(), lattice, ParticleSet::R, ParticleSet::resetGroups(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-B BCC H Ewald3D" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 94 of file test_coulomb_pbcAB_ewald.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAB::evalConsts(), CoulombPBCAA::evaluate(), CoulombPBCAB::evaluate(), ParticleSet::getSpeciesSet(), lattice, ParticleSet::R, ParticleSet::resetGroups(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "Crowd redistribute walkers" | ) |
Definition at line 94 of file test_Crowd.cpp.
References crowd, CrowdWithWalkers::get_crowd(), CrowdWithWalkers::hams, CrowdWithWalkers::makeAnotherPointWalker(), pools, CrowdWithWalkers::psets, REQUIRE(), CrowdWithWalkers::twfs, and CrowdWithWalkers::walkers.
| qmcplusplus::TEST_CASE | ( | "Gaussian Combo P" | , |
| "" | [numerics] | ||
| ) |
Definition at line 95 of file test_gaussian_basis.cpp.
References GaussianCombo< T >::addGaussian(), CHECK(), GaussianCombo< T >::d2Y, GaussianCombo< T >::d3Y, GaussianCombo< T >::df(), GaussianCombo< T >::dY, GaussianCombo< T >::evaluate(), GaussianCombo< T >::evaluateAll(), GaussianCombo< T >::evaluateWithThirdDeriv(), GaussianCombo< T >::f(), REQUIRE(), GaussianCombo< T >::size(), and GaussianCombo< T >::Y.
| qmcplusplus::TEST_CASE | ( | "InputSection::InputSection" | , |
| "" | [estimators] | ||
| ) |
Definition at line 96 of file test_InputSection.cpp.
References CHECK(), and InputSection::has().
| qmcplusplus::TEST_CASE | ( | "gaussian random input zero" | , |
| "" | [particle_base] | ||
| ) |
Definition at line 96 of file test_random_seq.cpp.
References assignGaussRand(), CHECK(), and FakeRandom< T >::set_value().
| qmcplusplus::TEST_CASE | ( | "ReadFileBuffer_sorep" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 96 of file test_ecp.cpp.
References CHECK(), OHMMS::Controller, getSplinedSOPot(), okay, ECPComponentBuilder::pp_so, ECPComponentBuilder::read_pp_file(), REQUIRE(), and ECPComponentBuilder::Zeff.
| qmcplusplus::TEST_CASE | ( | "fillOverlapAndHamiltonianMatrices" | , |
| "" | [drivers] | ||
| ) |
Definition at line 97 of file test_QMCCostFunctionBatched.cpp.
References CHECK(), comm, OHMMS::Controller, LinearMethodTestSupport::costFn, QMCCostFunctionBase::ENERGY_NEW, QMCCostFunctionBatched::fillOverlapHamiltonianMatrices(), LinearMethodTestSupport::getDerivRecords(), LinearMethodTestSupport::getHDerivRecords(), LinearMethodTestSupport::getRecordsOnNode(), LinearMethodTestSupport::getSumValue(), ham, qmcplusplus::Units::force::N, QMCCostFunctionBase::REWEIGHT, LinearMethodTestSupport::set_samples_and_param(), QMCCostFunctionBase::SUM_E_WGT, QMCCostFunctionBase::SUM_ESQ_WGT, and QMCCostFunctionBase::SUM_WGT.
| qmcplusplus::TEST_CASE | ( | "get scaled drift real" | , |
| "" | [drivers][drift] | ||
| ) |
Definition at line 97 of file test_drift.cpp.
References CHECK(), ParticleSet::create(), ParticleSet::G, DriftModifierUNR::getDrift(), ParticleSet::setName(), and sqrt().
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A Ewald2D triangular" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 97 of file test_ewald2d.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAA::evaluate(), ParticleSet::getSpeciesSet(), lattice, ParticleSet::R, ParticleSet::setName(), sqrt(), LRCoulombSingleton::STRICT2D, LRCoulombSingleton::this_lr_type, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "checkMatrix_real" | , |
| "" | [utilities][for_testing] | ||
| ) |
Definition at line 97 of file test_checkMatrix.cpp.
References CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "find minimum" | , |
| "" | [numerics] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "EstimatorManagerInput::moveFromEstimatorInputs" | , |
| "" | [estimators] | ||
| ) |
Definition at line 98 of file test_EstimatorManagerInput.cpp.
References doc, emi(), EstimatorManagerInput::get_estimator_inputs(), Libxml2Document::getRoot(), node, okay, Libxml2Document::parseFromString(), REQUIRE(), EstimatorManagerInputTests::testAppendFromXML(), and ValidSpinDensityInput::xml.
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A elec Ewald3D" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 98 of file test_coulomb_pbcAA_ewald.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAA::evalConsts(), CoulombPBCAA::evaluate(), ParticleSet::getSpeciesSet(), lattice, ParticleSet::R, ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "WalkerControl::determineNewWalkerPopulation" | , |
| "" | [drivers][walker_control] | ||
| ) |
Definition at line 99 of file test_WalkerControl.cpp.
References CHECK(), and UnifiedDriverWalkerControlMPITest::testNewDistribution().
| qmcplusplus::TEST_CASE | ( | "readLine" | , |
| "" | [utilities] | ||
| ) |
Definition at line 99 of file test_parser.cpp.
References readLine(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "NestedContainers" | , |
| "" | [OhmmsPETE] | ||
| ) |
Definition at line 100 of file test_Vector.cpp.
References CHECK(), Vector< T, Alloc >::clear(), REQUIRE(), Vector< T, Alloc >::resize(), and Vector< T, Alloc >::size().
| qmcplusplus::TEST_CASE | ( | "VectorSoaContainer assignment" | , |
| "" | [OhmmsSoA] | ||
| ) |
Definition at line 101 of file test_vector_soa.cpp.
References CHECK(), and VectorSoaContainer< T, D, Alloc >::copyIn().
| qmcplusplus::TEST_CASE | ( | "test_communicate_split_four" | , |
| "" | [message] | ||
| ) |
Definition at line 103 of file test_communciate.cpp.
References OHMMS::Controller, REQUIRE(), and Communicate::size().
| qmcplusplus::TEST_CASE | ( | "fairDivide_one" | , |
| "" | [utilities] | ||
| ) |
Definition at line 107 of file test_partition.cpp.
References fairDivide(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "Derivatives of Spherical Harmonics" | , |
| "" | [numerics] | ||
| ) |
Definition at line 107 of file test_ylm.cpp.
References CHECK(), derivYlmSpherical(), imag(), qmcplusplus::Units::distance::m, and qmcplusplus::Units::force::N.
| qmcplusplus::TEST_CASE | ( | "MomentumDistribution::accumulate" | , |
| "" | [estimators] | ||
| ) |
Definition at line 107 of file test_MomentumDistribution.cpp.
References ProjectData::BATCH, CHECK(), comm, OHMMS::Controller, convertUPtrToRefVector(), crowd, doc, qmcplusplus::Units::charge::e, Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), lattice, MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), qmcplusplus::testing::makeTestLattice(), node, okay, outputManager, Libxml2Document::parseFromString(), particle_pool, OutputManagerClass::pause(), pset, OutputManagerClass::resume(), test_project, walker, qmcplusplus::hdf::walkers, and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "walker buffer | add, |
| update | , | ||
| restore" | , | ||
| "" | [particle] | ||
| ) |
Definition at line 107 of file test_walker.cpp.
References CHECK(), and qmcplusplus::hdf::walkers.
| qmcplusplus::TEST_CASE | ( | "OutputManager pause" | , |
| "" | [utilities] | ||
| ) |
Definition at line 108 of file test_output_manager.cpp.
References app_log(), app_out, app_summary(), init_string_output(), outputManager, OutputManagerClass::pause(), REQUIRE(), reset_string_output(), OutputManagerClass::resume(), and summary_out.
| qmcplusplus::TEST_CASE | ( | "test_timer_flat_profile_same_name" | , |
| "" | [utilities] | ||
| ) |
Definition at line 109 of file test_timer.cpp.
References TimerManager< TIMER >::FlatProfileData::callList, CHECK(), TimerManager< TIMER >::collate_flat_profile(), convert_to_ns(), TimerManager< TIMER >::createTimer(), TimerManager< TIMER >::FlatProfileData::nameList, REQUIRE(), qmcplusplus::Units::time::s, TimerManager< TIMER >::set_timer_threshold(), TimerType< CLOCK >::start(), TimerType< CLOCK >::stop(), TimerManager< TIMER >::FlatProfileData::timeList, and timer_level_fine.
| qmcplusplus::TEST_CASE | ( | "ModernStringUtils_strip" | , |
| "" | [utilities] | ||
| ) |
Definition at line 109 of file test_ModernStringUtils.cpp.
References CHECK(), strip(), and qmcplusplus::modernstrutil::strip().
| qmcplusplus::TEST_CASE | ( | "accumulator with weights float" | , |
| "" | [estimators] | ||
| ) |
Definition at line 110 of file test_accumulator.cpp.
| qmcplusplus::TEST_CASE | ( | "TrialWaveFunction flex_evaluateParameterDerivatives" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 111 of file test_TrialWaveFunction_He.cpp.
References CHECK(), TrialWaveFunction::checkInVariables(), OHMMS::Controller, TrialWaveFunction::createResource(), TrialWaveFunction::evaluateDerivatives(), TrialWaveFunction::mw_evaluateParameterDerivatives(), ParticleSet::R, setup_He_wavefunction(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "InputSection::assignAnyEnum" | , |
| "" | [estimators] | ||
| ) |
Definition at line 112 of file test_InputSection.cpp.
References InputSection::get(), VALUE1, and VALUE2.
| qmcplusplus::TEST_CASE | ( | "cuBLAS_LU::computeLogDet_complex" | , |
| "" | [wavefunction][CUDA] | ||
| ) |
Definition at line 112 of file test_cuBLAS_LU.cpp.
References batch_size, CHECK(), qmcplusplus::cuBLAS_LU::computeLogDet_batched(), cudaErrorCheck(), cudaMemcpyAsync, cudaMemcpyDeviceToHost, cudaMemcpyHostToDevice, cudaStreamSynchronize, dev_log_values(), dev_lu(), dev_lus(), dev_pivots(), hstream, lda, log_values(), lu, lus, n, and pivots.
| qmcplusplus::TEST_CASE | ( | "fccz sr lr clone" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 112 of file test_force_ewald.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), clone, LRCoulombSingleton::CoulombDerivHandler, LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), ForceChiesaPBCAA::evaluateLR(), ForceChiesaPBCAA::evaluateSR(), ForceBase::getAddIonIon(), ForceBase::getForces(), ParticleSet::getSpeciesSet(), ForceChiesaPBCAA::InitMatrix(), lattice, ForceChiesaPBCAA::makeClone(), ParticleSet::R, REQUIRE(), ParticleSet::resetGroups(), ForceBase::setAddIonIon(), ForceBase::setForces(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "accumulator with weights double" | , |
| "" | [estimators] | ||
| ) |
Definition at line 112 of file test_accumulator.cpp.
| qmcplusplus::TEST_CASE | ( | "DiracMatrixComputeOMPTarget_large_determinants_against_legacy" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 113 of file test_DiracMatrixComputeOMPTarget.cpp.
References check_matrix_result, CHECKED_ELSE(), checkMatrix(), DiracMatrix< T_FP >::invert_transpose(), log_values(), qmcplusplus::testing::makeRngSpdMatrix(), DiracMatrixComputeOMPTarget< VALUE_FP >::mw_invertTranspose(), n, queue, and Matrix< T, Alloc >::resize().
| qmcplusplus::TEST_CASE | ( | "SPO input spline from xml He_sto3g" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 113 of file test_spo_collection_input_LCAO_xml.cpp.
References app_log(), and test_He_sto3g_xml_input().
| qmcplusplus::TEST_CASE | ( | "fairDivide_empties" | , |
| "" | [utilities] | ||
| ) |
Definition at line 114 of file test_partition.cpp.
References fairDivide(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "OhmmsMatrix_VectorSoaContainer_View" | , |
| "" | [Integration][Allocators] | ||
| ) |
Definition at line 114 of file test_dual_allocators_ohmms_containers.cpp.
| qmcplusplus::TEST_CASE | ( | "SPO input spline from HDF diamond_2x1x1" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 115 of file test_spo_collection_input_spline.cpp.
References app_log(), and test_diamond_2x1x1_xml_input().
| qmcplusplus::TEST_CASE | ( | "one_dim_cubic_spline_1" | , |
| "" | [numerics] | ||
| ) |
Definition at line 115 of file test_one_dim_cubic_spline.cpp.
References CHECK(), n, OneDimCubicSpline< Td, Tg, CTd, CTg >::spline(), and OneDimCubicSpline< Td, Tg, CTd, CTg >::splint().
| qmcplusplus::TEST_CASE | ( | "J1 evaluate derivatives Jastrow with two species" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 115 of file test_J1OrbitalSoA.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), ParticleSet::addTable(), app_log(), WaveFunctionFactory::buildTWF(), CHECK(), OHMMS::Controller, ParticleSet::create(), doc, ParticleSet::get(), ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), ispecies, okay, Libxml2Document::parseFromString(), ParticleSet::R, VariableSet::removeInactive(), REQUIRE(), ParticleSet::resetGroups(), ParticleSet::setName(), VariableSet::size_of_active(), twf, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "EstimatorManagerNew::collectScalarEstimators" | , |
| "" | [estimators] | ||
| ) |
Definition at line 116 of file test_EstimatorManagerNew.cpp.
References CHECK(), OHMMS::Controller, EstimatorManagerNewTest::em, embt, EstimatorManagerNewTest::fakeScalarSamplesAndCollect(), EstimatorManagerNew::get_AverageCache(), and ham.
| qmcplusplus::TEST_CASE | ( | "Pade2 Jastrow" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 117 of file test_pade_jastrow.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), OHMMS::Controller, ParticleSet::create(), doc, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), ispecies, okay, Libxml2Document::parseFromString(), ParticleSet::R, VariableSet::removeInactive(), REQUIRE(), ParticleSet::resetGroups(), ParticleSet::setName(), VariableSet::size_of_active(), twf, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "Walker control assign walkers odd ranks" | , |
| "" | [drivers][walker_control] | ||
| ) |
Definition at line 118 of file test_walker_control.cpp.
References WalkerControlMPI::determineNewWalkerPopulation(), qmcplusplus::Units::mass::me, and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "SpaceGrid::Construction" | , |
| "" | [estimators] | ||
| ) |
Definition at line 119 of file test_NESpaceGrid.cpp.
References CHECK(), comm, OHMMS::Controller, OHMMS_DIM, SpaceGridEnv< VALID >::ref_points_, and SpaceGridEnv< VALID >::sgi_.
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A elec" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 119 of file test_coulomb_pbcAA.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAA::evalConsts(), CoulombPBCAA::evaluate(), ParticleSet::getSpeciesSet(), lattice, ParticleSet::R, ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "benchmark_DiracMatrixComputeCUDA_vs_legacy_256_10" | , |
| "" | [wavefunction][fermion][benchmark] | ||
| ) |
This test will run by default.
Definition at line 120 of file benchmark_DiracMatrixComputeCUDA.cpp.
References DiracComputeBenchmarkParameters::batch_size, DiracMatrix< T_FP >::invert_transpose(), log_values(), qmcplusplus::testing::makeRngSpdMatrix(), qmcplusplus::Units::meter, DiracMatrixComputeCUDA< VALUE_FP >::mw_invertTranspose(), DiracComputeBenchmarkParameters::n, DiracComputeBenchmarkParameters::name, queue, and DiracComputeBenchmarkParameters::str().
| qmcplusplus::TEST_CASE | ( | "read_lattice_xml_lrhandle" | , |
| "" | [particle_io][xml] | ||
| ) |
Definition at line 121 of file test_lattice_parser.cpp.
References CHECK(), doc, Libxml2Document::getRoot(), okay, Libxml2Document::parseFromString(), LatticeParser::put(), CrystalLattice< T, D >::R, and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "particle set lattice with vacuum" | , |
| "" | [particle] | ||
| ) |
Definition at line 122 of file test_ParticleSet.cpp.
References CrystalLattice< T, D >::BoxBConds, CHECK(), ParticleSet::createSK(), ParticleSet::getLRBox(), CrystalLattice< T, D >::R, CrystalLattice< T, D >::reset(), ParticleSet::setName(), SUPERCELL_BULK, SUPERCELL_SLAB, SUPERCELL_WIRE, CrystalLattice< T, D >::SuperCellEnum, and CrystalLattice< T, D >::VacuumScale.
| qmcplusplus::TEST_CASE | ( | "fairDivide_typical" | , |
| "" | [utilities] | ||
| ) |
Definition at line 123 of file test_partition.cpp.
References fairDivide(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "InputSection::readXML" | , |
| "" | [estimators] | ||
| ) |
Definition at line 124 of file test_InputSection.cpp.
References CHECK(), doc, InputSection::get(), Libxml2Document::getRoot(), InputSection::has(), okay, Libxml2Document::parseFromString(), InputSection::readXML(), TestInputSection::report(), REQUIRE(), VALUE1, and VALUE2.
| qmcplusplus::TEST_CASE | ( | "kcontainer at twist in 2D" | , |
| "" | [longrange] | ||
| ) |
Definition at line 124 of file test_kcontainer.cpp.
References CHECK(), qmcplusplus::Units::charge::e, KContainer::kpts, KContainer::kpts_cart, lattice, and KContainer::updateKLists().
| qmcplusplus::TEST_CASE | ( | "SODMC" | , |
| "" | [drivers][dmc] | ||
| ) |
Definition at line 125 of file test_dmc_driver.cpp.
References CloneManager::acceptRatio(), SpeciesSet::addAttribute(), TrialWaveFunction::addComponent(), QMCHamiltonian::addOperator(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), CloneManager::clearClones(), OHMMS::Controller, ParticleSet::create(), MCWalkerConfiguration::createWalkers(), doc, Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), ParticleSet::getSpeciesSet(), okay, omp_get_max_threads(), Libxml2Document::parseFromString(), QMCDriver::process(), ParticleSet::R, REQUIRE(), DMC::run(), ParticleSet::setName(), ParticleSet::setSpinor(), ParticleSet::spins, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "SOVMC" | , |
| "" | [drivers][vmc] | ||
| ) |
Definition at line 125 of file test_vmc_driver.cpp.
References CloneManager::acceptRatio(), SpeciesSet::addAttribute(), TrialWaveFunction::addComponent(), QMCHamiltonian::addOperator(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), CloneManager::clearClones(), OHMMS::Controller, ParticleSet::create(), MCWalkerConfiguration::createWalkers(), doc, Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), ParticleSet::getSpeciesSet(), okay, omp_get_max_threads(), Libxml2Document::parseFromString(), QMCDriver::process(), ParticleSet::R, REQUIRE(), MCWalkerConfiguration::resetWalkerProperty(), VMC::run(), Communicate::setName(), ParticleSet::setName(), ParticleSet::setSpinor(), ParticleSet::spins, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A Ewald Quasi2D staggered square" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 126 of file test_ewald_quasi2d.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAA::evaluate(), ParticleSet::getSpeciesSet(), lattice, LRCoulombSingleton::QUASI2D, ParticleSet::R, ParticleSet::setName(), LRCoulombSingleton::this_lr_type, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "DiracDeterminantBatched_first" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 126 of file test_DiracDeterminantBatched.cpp.
| qmcplusplus::TEST_CASE | ( | "Cartesian Tensor evaluateAll subset" | , |
| "" | [numerics] | ||
| ) |
Definition at line 126 of file test_cartesian_tensor.cpp.
References CHECK(), CartesianTensor< T, Point_t, Tensor_t, GGG_t >::evaluateAll(), CartesianTensor< T, Point_t, Tensor_t, GGG_t >::getGradYlm(), CartesianTensor< T, Point_t, Tensor_t, GGG_t >::getLaplYlm(), and CartesianTensor< T, Point_t, Tensor_t, GGG_t >::getYlm().
| qmcplusplus::TEST_CASE | ( | "distance_open_xy" | , |
| "" | [distance_table][xml] | ||
| ) |
Definition at line 126 of file test_distance_table.cpp.
References ParticleSet::addTable(), CHECK(), doc, ParticleSet::getDistTableAB(), OhmmsElementBase::getName(), Libxml2Document::getRoot(), ParticleSet::getTotalNum(), ParticleSet::isSameMass(), okay, Libxml2Document::parseFromString(), XMLParticleParser::readXML(), REQUIRE(), sqrt(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "DMC Particle-by-Particle advanceWalkers LinearOrbital" | , |
| "" | [drivers][dmc] | ||
| ) |
Definition at line 127 of file test_dmc.cpp.
References SpeciesSet::addAttribute(), TrialWaveFunction::addComponent(), QMCHamiltonian::addObservables(), QMCHamiltonian::addOperator(), SpeciesSet::addSpecies(), ParticleSet::addTable(), QMCUpdateBase::advanceWalkers(), WalkerConfigurations::begin(), CHECK(), ParticleSet::create(), MCWalkerConfiguration::createWalkers(), WalkerConfigurations::end(), ParticleSet::getSpeciesSet(), QMCUpdateBase::nAccept, QMCUpdateBase::nReject, ParticleSet::R, TrialWaveFunction::registerData(), REQUIRE(), QMCUpdateBase::resetRun(), MCWalkerConfiguration::resetWalkerProperty(), ParticleSet::setName(), QMCUpdateBase::startBlock(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "CountingJastrow" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 128 of file test_counting_jastrow.cpp.
References CountingJastrow< RegionType >::acceptMove(), Vector< T, Alloc >::begin(), CountingJastrowBuilder::buildComponent(), CHECK(), CountingJastrow< RegionType >::checkInVariablesExclusive(), CountingJastrow< RegionType >::checkOutVariables(), OHMMS::Controller, ParticleSet::create(), doc, qmcplusplus::Units::charge::e, Vector< T, Alloc >::end(), CountingJastrow< RegionType >::evalGrad(), CountingJastrow< RegionType >::evaluateDerivatives(), CountingJastrow< RegionType >::evaluateLog(), ParticleSet::G, WaveFunctionComponent::get_log_value(), Libxml2Document::getRoot(), ParticleSet::L, CountingJastrow< RegionType >::makeClone(), ParticleSet::makeMoveAndCheck(), Libxml2Document::parseFromString(), ParticleSet::R, CountingJastrow< RegionType >::ratio(), CountingJastrow< RegionType >::ratioGrad(), CountingJastrow< RegionType >::recompute(), REQUIRE(), CountingJastrow< RegionType >::resetParametersExclusive(), Vector< T, Alloc >::resize(), and ParticleSet::setName().
| qmcplusplus::TEST_CASE | ( | "EstimatorManagerInput::moveConstructor" | , |
| "" | [estimators] | ||
| ) |
Definition at line 128 of file test_EstimatorManagerInput.cpp.
References CHECK(), qmcplusplus::testing::createEstimatorManagerNewInputXML(), emi(), estimators_doc, EstimatorManagerInput::get_estimator_inputs(), EstimatorManagerInput::get_scalar_estimator_inputs(), and Libxml2Document::getRoot().
| qmcplusplus::TEST_CASE | ( | "Gaussian Combo D" | , |
| "" | [numerics] | ||
| ) |
Definition at line 129 of file test_gaussian_basis.cpp.
References GaussianCombo< T >::addGaussian(), CHECK(), GaussianCombo< T >::d2Y, GaussianCombo< T >::d3Y, GaussianCombo< T >::df(), GaussianCombo< T >::dY, GaussianCombo< T >::evaluate(), GaussianCombo< T >::evaluateAll(), GaussianCombo< T >::evaluateWithThirdDeriv(), GaussianCombo< T >::f(), REQUIRE(), GaussianCombo< T >::size(), and GaussianCombo< T >::Y.
| qmcplusplus::TEST_CASE | ( | "SoA Cartesian Tensor evaluateVGL subset" | , |
| "" | [numerics] | ||
| ) |
Definition at line 129 of file test_soa_cartesian_tensor.cpp.
References CHECK(), SoaCartesianTensor< T >::evaluateVGL(), and SoaCartesianTensor< T >::getcXYZ().
| qmcplusplus::TEST_CASE | ( | "Chiesa Force" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 130 of file test_force.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), check_force_copy(), ParticleSet::create(), ParticleSet::createSK(), ForceChiesaPBCAA::evaluate(), CoulombPBCAA::evaluateWithIonDerivs(), LRCoulombSingleton::EWALD, ForceBase::getForces(), ForceBase::getForcesIonIon(), ParticleSet::getSpeciesSet(), ForceChiesaPBCAA::InitMatrix(), lattice, ForceChiesaPBCAA::makeClone(), ParticleSet::R, REQUIRE(), ParticleSet::resetGroups(), ForceBase::setAddIonIon(), ParticleSet::setName(), LRCoulombSingleton::this_lr_type, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "OutputManager shutoff" | , |
| "" | [utilities] | ||
| ) |
Definition at line 131 of file test_output_manager.cpp.
References app_debug, app_error(), app_log(), app_out, app_summary(), debug_out, err_out, init_string_output(), outputManager, REQUIRE(), OutputManagerClass::shutOff(), and summary_out.
| qmcplusplus::TEST_CASE | ( | "J1 spin evaluate derivatives multiparticle Jastrow" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 132 of file test_J1Spin.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), OHMMS::Controller, ParticleSet::create(), doc, ParticleSet::G, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), ispecies, ParticleSet::L, log(), okay, Libxml2Document::parseFromString(), ParticleSet::R, VariableSet::removeInactive(), REQUIRE(), ParticleSet::resetGroups(), ParticleSet::setName(), VariableSet::size_of_active(), twf, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "ReadMolecularOrbital GTO He" | , |
| "" | [wavefunction] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "ReadFileBuffer_reopen" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 132 of file test_ecp.cpp.
References ReadFileBuffer::length, ReadFileBuffer::open_file(), ReadFileBuffer::read_contents(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "fairDivide_even" | , |
| "" | [utilities] | ||
| ) |
Definition at line 132 of file test_partition.cpp.
References fairDivide(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "ParticleSetPool putLattice" | , |
| "" | [qmcapp] | ||
| ) |
Definition at line 132 of file test_particle_pool.cpp.
References OHMMS::Controller, doc, Libxml2Document::getRoot(), lattice, okay, Libxml2Document::parseFromString(), ParticleSetPool::readSimulationCellXML(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "PlaneWave SPO from HDF for LiH arb" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 133 of file test_pw.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), OHMMS::Controller, ParticleSet::create(), SPOSetBuilder::createSPOSet(), doc, Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), imag(), lattice, okay, Libxml2Document::parseFromString(), ParticleSet::R, real(), REQUIRE(), ParticleSet::resetGroups(), ParticleSet::setName(), ParticleSetPool::setSimulationCell(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "ReadMolecularOrbital Numerical He" | , |
| "" | [wavefunction] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "DiracMatrixComputeCUDA_complex_determinants_against_legacy" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 134 of file test_DiracMatrixComputeCUDA.cpp.
References check_matrix_result, CHECKED_ELSE(), checkMatrix(), DiracMatrix< T_FP >::invert_transpose(), log_values(), qmcplusplus::testing::makeRngSpdMatrix(), DiracMatrixComputeCUDA< VALUE_FP >::mw_invertTranspose(), n, queue, and Matrix< T, Alloc >::resize().
| qmcplusplus::TEST_CASE | ( | "DiracMatrix_inverse_matching_2" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 137 of file test_DiracMatrix.cpp.
References CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), Matrix< T, Alloc >::cols(), Matrix< T, Alloc >::data(), DiracMatrix< T_FP >::invert_transpose(), lu, lu_mat(), Matrix< T, Alloc >::resize(), Matrix< T, Alloc >::rows(), qmcplusplus::simd::transpose(), and Xgetrf().
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A Ewald2D honeycomb" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 137 of file test_ewald2d.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), dot(), CoulombPBCAA::evaluate(), ParticleSet::getSpeciesSet(), lattice, ParticleSet::R, ParticleSet::setName(), sqrt(), LRCoulombSingleton::STRICT2D, LRCoulombSingleton::this_lr_type, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "EstimatorManagerNew adhoc addVector operator" | , |
| "" | [estimators] | ||
| ) |
Definition at line 137 of file test_EstimatorManagerNew.cpp.
References REQUIRE().
Definition at line 138 of file test_QMCDriverFactory.cpp.
References CHECK(), comm, OHMMS::Controller, qmcplusplus::testing::createDriver(), DMC, doc, Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), QMCDriverFactory::DriverAssemblyState::new_run_type, node, okay, Libxml2Document::parseFromString(), QMCDriverFactory::readSection(), REQUIRE(), test_project, qmcplusplus::testing::valid_dmc_input_dmc_index, and qmcplusplus::testing::valid_dmc_input_sections.
| qmcplusplus::TEST_CASE | ( | "getwords" | , |
| "" | [utilities] | ||
| ) |
Definition at line 138 of file test_parser.cpp.
References getwords(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "MCPopulation::redistributeWalkers" | , |
| "" | [particle][population] | ||
| ) |
Definition at line 139 of file test_MCPopulation.cpp.
References comm, OHMMS::Controller, MCPopulation::createWalkers(), hamiltonian_pool, MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), MinimalHamiltonianPool::make_hamWithEE(), particle_pool, Communicate::rank(), REQUIRE(), qmcplusplus::hdf::walkers, and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "DiracDeterminant_first" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 142 of file test_DiracDeterminant.cpp.
| qmcplusplus::TEST_CASE | ( | "read_dynamic_spin_eset_xml" | , |
| "" | [particle_io][xml] | ||
| ) |
Definition at line 142 of file test_xml_io.cpp.
References CHECK(), doc, OhmmsElementBase::getName(), Libxml2Document::getRoot(), ParticleSet::groups(), okay, Libxml2Document::parseFromString(), ParticleSet::R, XMLParticleParser::readXML(), REQUIRE(), Vector< T, Alloc >::size(), and ParticleSet::spins.
| qmcplusplus::TEST_CASE | ( | "EstimatorManagerInput::MergeConstructor" | , |
| "" | [estimators] | ||
| ) |
Definition at line 143 of file test_EstimatorManagerInput.cpp.
References CHECK(), qmcplusplus::testing::createEstimatorManagerNewGlobalInputXML(), qmcplusplus::testing::createEstimatorManagerNewInputXML(), estimators_doc, and Libxml2Document::getRoot().
| qmcplusplus::TEST_CASE | ( | "SpaceGrid::CYLINDRICAL" | , |
| "" | [estimators] | ||
| ) |
Definition at line 144 of file test_NESpaceGrid.cpp.
References CHECK(), comm, OHMMS::Controller, OHMMS_DIM, SpaceGridEnv< VALID >::ref_points_, and SpaceGridEnv< VALID >::sgi_.
| qmcplusplus::TEST_CASE | ( | "ompBLAS gemm" | , |
| "" | [OMP] | ||
| ) |
Definition at line 146 of file test_ompBLAS.cpp.
References qmcplusplus::Units::energy::K, and qmcplusplus::Units::force::N.
| qmcplusplus::TEST_CASE | ( | "TwoBodyJastrow variables fail" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 149 of file test_J2_derivatives.cpp.
References TwoBodyJastrow< FT >::addFunc(), CHECK(), TwoBodyJastrow< FT >::checkOutVariables(), TwoBodyJastrow< FT >::evaluateDerivatives(), TwoBodyJastrow< FT >::evaluateDerivativesWF(), get_two_species_particleset(), TwoBodyJastrow< FT >::getComponentOffset(), VariableSet::insertFrom(), VariableSet::resetIndex(), and VariableSet::size_of_active().
| qmcplusplus::TEST_CASE | ( | "test_timer_nested_profile" | , |
| "" | [utilities] | ||
| ) |
Definition at line 150 of file test_timer.cpp.
References CHECK(), TimerManager< TIMER >::collate_flat_profile(), TimerManager< TIMER >::collate_stack_profile(), convert_to_ns(), convert_to_s(), TimerManager< TIMER >::createTimer(), TimerManager< TIMER >::FlatProfileData::nameList, TimerManager< TIMER >::StackProfileData::nameList, REQUIRE(), qmcplusplus::Units::time::s, TimerManager< TIMER >::set_timer_threshold(), TimerType< CLOCK >::start(), TimerType< CLOCK >::stop(), TimerManager< TIMER >::StackProfileData::timeExclList, TimerManager< TIMER >::FlatProfileData::timeList, TimerManager< TIMER >::StackProfileData::timeList, and timer_level_fine.
| qmcplusplus::TEST_CASE | ( | "QMCDriverNew test driver operations" | , |
| "" | [drivers] | ||
| ) |
Definition at line 150 of file test_QMCDriverNew.cpp.
References ProjectData::BATCH, CHECK(), comm, QMCDriverNew::computeLogGreensFunction(), OHMMS::Controller, doc, Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), hamiltonian_pool, MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), MinimalHamiltonianPool::make_hamWithEE(), node, okay, outputManager, Libxml2Document::parseFromString(), particle_pool, OutputManagerClass::pause(), POS, POS_SPIN, Communicate::rank(), QMCDriverInput::readXML(), REQUIRE(), OutputManagerClass::resume(), QMCDriverNew::scaleBySqrtTau(), Communicate::size(), test_project, QMCDriverNewTestWrapper::testDetermineStepsPerBlock(), QMCDriverNewTestWrapper::testMeasureImbalance(), qmcplusplus::testing::valid_vmc_input_sections, qmcplusplus::testing::valid_vmc_input_vmc_tiny_index, and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "ProjectData::TestIsComplex" | , |
| "" | [ohmmsapp] | ||
| ) |
Definition at line 153 of file test_project_data.cpp.
References ProjectData::isComplex(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "SlaterDet mw_ APIs" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 153 of file test_SlaterDet.cpp.
References CHECK(), comm, OHMMS::Controller, SlaterDet::createResource(), MinimalParticlePool::make_O2_spinor(), SlaterDet::mw_evalGrad(), SlaterDet::mw_evalGradWithSpin(), SlaterDet::mw_ratioGrad(), SlaterDet::mw_ratioGradWithSpin(), and particle_pool.
| qmcplusplus::TEST_CASE | ( | "CoulombAB::Listener" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 154 of file test_coulomb_pbcAB.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), Matrix< T, Alloc >::begin(), CHECK(), Matrix< T, Alloc >::cols(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), CoulombPBCAB::createResource(), ParticleSet::createResource(), ParticleSet::createSK(), CoulombPBCAB::evalConsts(), qmcplusplus::testing::getParticularListener(), ParticleSet::getSpeciesSet(), OperatorBase::getValue(), lattice, CoulombPBCAB::mw_evaluatePerParticle(), ParticleSet::mw_update(), ParticleSet::R, ParticleSet::setName(), ParticleSet::turnOnPerParticleSK(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A BCC" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 155 of file test_coulomb_pbcAA.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAA::evaluate(), CoulombPBCAA::get_madelung_constant(), ParticleSet::getSpeciesSet(), lattice, ParticleSet::R, ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "Evaluate_ecp" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 156 of file test_ecp.cpp.
References SpeciesSet::addAttribute(), TrialWaveFunction::addComponent(), SpeciesSet::addSpecies(), ParticleSet::addTable(), RadialJastrowBuilder::buildComponent(), CHECK(), TrialWaveFunction::checkInVariables(), TrialWaveFunction::checkOutVariables(), OHMMS::Controller, copyGridUnrotatedForTest(), ParticleSet::create(), ParticleSet::createSK(), NonLocalECPComponent::deleteVirtualParticle(), doc, qmcplusplus::Units::charge::e, TrialWaveFunction::evaluateDerivatives(), TrialWaveFunction::evaluateLog(), NonLocalECPComponent::evaluateOne(), NonLocalECPComponent::evaluateOneWithForces(), NonLocalECPComponent::evaluateValueAndDerivatives(), ParticleSet::getDistTableAB(), NonLocalECPComponent::getRmax(), Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), ParticleSet::getTotalNum(), NonLocalECPComponent::initVirtualParticle(), lattice, okay, Libxml2Document::parseFromString(), ECPComponentBuilder::pp_nonloc, ParticleSet::R, ECPComponentBuilder::read_pp_file(), REQUIRE(), ParticleSet::resetGroups(), VariableSet::resetIndex(), Vector< T, Alloc >::resize(), ParticleSet::setName(), VariableSet::size(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "Spherical Harmonics Wrapper" | , |
| "" | [numerics] | ||
| ) |
Definition at line 162 of file test_ylm.cpp.
References CHECK(), qmcplusplus::Units::distance::m, qmcplusplus::Units::force::N, and sphericalHarmonic().
| qmcplusplus::TEST_CASE | ( | "QMCDriverFactory create DMCBatched driver" | , |
| "" | [qmcapp] | ||
| ) |
Definition at line 162 of file test_QMCDriverFactory.cpp.
References ProjectData::BATCH, CHECK(), comm, OHMMS::Controller, qmcplusplus::testing::createDriver(), DMC_BATCH, doc, Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), QMCDriverFactory::DriverAssemblyState::new_run_type, node, okay, Libxml2Document::parseFromString(), QMCDriverFactory::readSection(), REQUIRE(), test_project, qmcplusplus::testing::valid_dmc_batch_input_dmc_batch_index, qmcplusplus::testing::valid_dmc_input_dmc_batch_index, and qmcplusplus::testing::valid_dmc_input_sections.
| qmcplusplus::TEST_CASE | ( | "Eloc_Derivatives:slater_noj" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 163 of file test_ion_derivs.cpp.
References app_log(), WaveFunctionFactory::buildTWF(), CHECK(), OHMMS::Controller, convertToReal(), create_CN_Hamiltonian(), create_CN_particlesets(), dot(), qmcplusplus::Units::charge::e, TrialWaveFunction::evalGradSource(), QMCHamiltonian::evaluateDeterministic(), QMCHamiltonian::evaluateIonDerivs(), QMCHamiltonian::evaluateIonDerivsDeterministic(), TrialWaveFunction::evaluateLog(), QMCHamiltonian::getHamiltonian(), QMCHamiltonian::getObservable(), QMCHamiltonian::getObservableName(), Libxml2Document::getRoot(), ham, Libxml2Document::parse(), REQUIRE(), Vector< T, Alloc >::resize(), QMCHamiltonian::setRandomGenerator(), and QMCHamiltonian::sizeOfObservables().
| qmcplusplus::TEST_CASE | ( | "Cartesian Gaussian Ordering" | , |
| "" | [wavefunction] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "getwordsWithMergedNumbers" | , |
| "" | [utilities] | ||
| ) |
Definition at line 165 of file test_parser.cpp.
References getwordsWithMergedNumbers(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "ReferencePoints::DefaultConstruction" | , |
| "" | [estimators] | ||
| ) |
Definition at line 165 of file test_ReferencePoints.cpp.
References approxEquality(), CHECK(), expectedReferencePoints(), generate_test_data, NEReferencePoints::get_points(), makePsets(), and makeTestRPI().
| qmcplusplus::TEST_CASE | ( | "Dirac Cartesian Gaussian Ordering" | , |
| "" | [wavefunction] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "SpaceGrid::SPHERICAL" | , |
| "" | [estimators] | ||
| ) |
Definition at line 166 of file test_NESpaceGrid.cpp.
References CHECK(), comm, OHMMS::Controller, OHMMS_DIM, SpaceGridEnv< VALID >::ref_points_, and SpaceGridEnv< VALID >::sgi_.
| qmcplusplus::TEST_CASE | ( | "Spline applyRotation one rotation" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 167 of file test_spline_applyrotation.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), CHECK(), CHECKED_ELSE(), checkMatrix(), OHMMS::Controller, ParticleSet::create(), EinsplineSetBuilder::createSPOSetFromXML(), doc, qmcplusplus::Units::charge::e, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), lattice, norm(), okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), ParticleSet::setName(), ParticleSetPool::setSimulationCell(), Vector< T, Alloc >::size(), and sqrt().
| qmcplusplus::TEST_CASE | ( | "RandomForTest_fillBufferRng_real" | , |
| "" | [utilities][for_testing] | ||
| ) |
Definition at line 167 of file test_RandomForTest.cpp.
| qmcplusplus::TEST_CASE | ( | "cuBLAS_LU::computeLogDet_float" | , |
| "" | [wavefunction][CUDA] | ||
| ) |
while this working is a good test, in production code its likely we want to widen the matrix M to double and thereby the LU matrix as well.
Definition at line 171 of file test_cuBLAS_LU.cpp.
References batch_size, CHECK(), qmcplusplus::cuBLAS_LU::computeLogDet_batched(), cudaCheck, cudaMemcpyAsync, cudaMemcpyDeviceToHost, cudaMemcpyHostToDevice, cudaStreamSynchronize, dev_log_values(), dev_lu(), dev_lus(), dev_pivots(), hstream, lda, log_values(), lu, lus, n, and pivots.
| qmcplusplus::TEST_CASE | ( | "benchmark_DiracMatrixComputeCUDASingle_vs_legacy_256_10" | , |
| "" | [wavefunction][fermion][.benchmark] | ||
| ) |
Only runs if [benchmark] tag is passed.
Definition at line 173 of file benchmark_DiracMatrixComputeCUDA.cpp.
References DiracComputeBenchmarkParameters::batch_size, DiracMatrix< T_FP >::invert_transpose(), DiracMatrixComputeCUDA< VALUE_FP >::invert_transpose(), log_values(), qmcplusplus::testing::makeRngSpdMatrix(), qmcplusplus::Units::meter, DiracComputeBenchmarkParameters::n, DiracComputeBenchmarkParameters::name, queue, and DiracComputeBenchmarkParameters::str().
| qmcplusplus::TEST_CASE | ( | "RandomForTest_fillVectorRngReal" | , |
| "" | [utilities][for_testing] | ||
| ) |
Definition at line 175 of file test_RandomForTest.cpp.
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A Ewald Quasi2D staggered square 2x2" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 175 of file test_ewald_quasi2d.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAA::evaluate(), ParticleSet::getSpeciesSet(), lattice, LRCoulombSingleton::QUASI2D, ParticleSet::R, ParticleSet::setName(), LRCoulombSingleton::this_lr_type, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "Cartesian Tensor evaluateWithHessian subset" | , |
| "" | [numerics] | ||
| ) |
Definition at line 180 of file test_cartesian_tensor.cpp.
References CHECK(), CartesianTensor< T, Point_t, Tensor_t, GGG_t >::evaluateWithHessian(), CartesianTensor< T, Point_t, Tensor_t, GGG_t >::getGradYlm(), CartesianTensor< T, Point_t, Tensor_t, GGG_t >::getHessYlm(), and CartesianTensor< T, Point_t, Tensor_t, GGG_t >::getYlm().
| qmcplusplus::TEST_CASE | ( | "RandomForTest_getVecRngReal" | , |
| "" | [utilities][for_testing] | ||
| ) |
Definition at line 183 of file test_RandomForTest.cpp.
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A Ewald2D tri. in rect." | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 184 of file test_ewald2d.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), app_log(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), CoulombPBCAA::evaluate(), ParticleSet::getSpeciesSet(), ParticleSet::getTotalNum(), lattice, ParticleSet::R, ParticleSet::setName(), sqrt(), LRCoulombSingleton::STRICT2D, LRCoulombSingleton::this_lr_type, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "ReferencePoints::Construction" | , |
| "" | [estimators] | ||
| ) |
Definition at line 187 of file test_ReferencePoints.cpp.
References approxEquality(), CHECK(), expectedReferencePoints(), generate_test_data, NEReferencePoints::get_points(), makePsets(), and makeTestRPI().
| qmcplusplus::TEST_CASE | ( | "SpinDensityNew::accumulate" | , |
| "" | [estimators] | ||
| ) |
Definition at line 187 of file test_SpinDensityNew.cpp.
References SpinDensityNew::accumulate(), CHECK(), doc, OperatorEstBase::get_data(), Libxml2Document::getRoot(), ValidSpinDensityInput::GRID, iattribute, ispecies, node, okay, Libxml2Document::parseFromString(), pset, REQUIRE(), sdi, sdn, species_set, qmcplusplus::hdf::walkers, and ValidSpinDensityInput::xml.
| qmcplusplus::TEST_CASE | ( | "SoA Cartesian Tensor evaluateVGH subset" | , |
| "" | [numerics] | ||
| ) |
Definition at line 187 of file test_soa_cartesian_tensor.cpp.
References CHECK(), SoaCartesianTensor< T >::evaluateVGH(), and SoaCartesianTensor< T >::getcXYZ().
| qmcplusplus::TEST_CASE | ( | "SpaceGrid::Basic" | , |
| "" | [estimators] | ||
| ) |
Definition at line 188 of file test_NESpaceGrid.cpp.
References ParticleSet::addTable(), ParticleSet::applyMinimumImage(), CHECK(), comm, OHMMS::Controller, ParticleSet::getDistTableAB(), ParticleSet::getTotalNum(), OHMMS_DIM, SpaceGridEnv< VALID >::pset_elec_, SpaceGridEnv< VALID >::pset_ions_, ParticleSet::R, SpaceGridEnv< VALID >::ref_points_, Matrix< T, Alloc >::resize(), SpaceGridEnv< VALID >::sgi_, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "OmpBLAS gemv" | , |
| "" | [SYCL] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "DiracMatrixComputeCUDA_large_determinants_against_legacy" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 189 of file test_DiracMatrixComputeCUDA.cpp.
References check_matrix_result, CHECKED_ELSE(), checkMatrix(), DiracMatrix< T_FP >::invert_transpose(), log_values(), qmcplusplus::testing::makeRngSpdMatrix(), DiracMatrixComputeCUDA< VALUE_FP >::mw_invertTranspose(), n, queue, and Matrix< T, Alloc >::resize().
| qmcplusplus::TEST_CASE | ( | "Hybridrep SPO from HDF diamond_2x1x1" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 191 of file test_hybridrep.cpp.
References abs(), SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), app_log(), CHECK(), OHMMS::Controller, ParticleSet::create(), ParticleSet::createResource(), EinsplineSetBuilder::createSPOSetFromXML(), doc, dot(), qmcplusplus::Units::charge::e, ParticleSet::getDistTable(), ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), lattice, okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), ParticleSet::setName(), ParticleSetPool::setSimulationCell(), Vector< T, Alloc >::size(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "RandomForTest_fillBufferRngComplex" | , |
| "" | [utilities][for_testing] | ||
| ) |
Definition at line 191 of file test_RandomForTest.cpp.
| qmcplusplus::TEST_CASE | ( | "test_timer_nested_profile_two_children" | , |
| "" | [utilities] | ||
| ) |
Definition at line 193 of file test_timer.cpp.
References TimerManager< TIMER >::collate_stack_profile(), convert_to_ns(), TimerManager< TIMER >::createTimer(), doc, Libxml2Document::dump(), Libxml2Document::getRoot(), TimerManager< TIMER >::StackProfileData::names, Libxml2Document::newDoc(), TimerManager< TIMER >::output_timing(), REQUIRE(), qmcplusplus::Units::time::s, TimerManager< TIMER >::set_timer_threshold(), TimerType< CLOCK >::start(), TimerType< CLOCK >::stop(), and timer_level_fine.
| qmcplusplus::TEST_CASE | ( | "RandomForTest_fillVecRngComplex" | , |
| "" | [utilities][for_testing] | ||
| ) |
Definition at line 199 of file test_RandomForTest.cpp.
| qmcplusplus::TEST_CASE | ( | "fillfromText" | , |
| "" | [drivers] | ||
| ) |
Definition at line 199 of file test_QMCCostFunctionBatched.cpp.
References fill_from_text(), and get_diamond_fill_data().
| qmcplusplus::TEST_CASE | ( | "fccz h3" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 202 of file test_force_ewald.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), LRCoulombSingleton::CoulombDerivHandler, LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), ForceChiesaPBCAA::evaluate(), ForceBase::getForces(), ForceBase::getForcesIonIon(), ParticleSet::getSpeciesSet(), ForceChiesaPBCAA::InitMatrix(), lattice, ParticleSet::R, ParticleSet::resetGroups(), ForceBase::setAddIonIon(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "Pair Correlation Pair Index" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 204 of file test_PairCorrEstimator.cpp.
References PairCorrEstimator::gen_pair_id(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "RandomForTest_getVecRngComplex" | , |
| "" | [utilities][for_testing] | ||
| ) |
Definition at line 207 of file test_RandomForTest.cpp.
| qmcplusplus::TEST_CASE | ( | "ReferencePoints::Description" | , |
| "" | [estimators] | ||
| ) |
Definition at line 209 of file test_ReferencePoints.cpp.
References CHECK(), makePsets(), makeTestRPI(), and NEReferencePoints::write_description().
| qmcplusplus::TEST_CASE | ( | "Cartesian derivatives of Spherical Harmonics" | , |
| "" | [numerics] | ||
| ) |
Definition at line 212 of file test_ylm.cpp.
References CHECK(), imag(), qmcplusplus::Units::distance::m, qmcplusplus::Units::force::N, and sphericalHarmonicGrad().
| qmcplusplus::TEST_CASE | ( | "OmpBLAS gemv notrans" | , |
| "" | [SYCL] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "distance_open_species_deviation" | , |
| "" | [distance_table][xml] | ||
| ) |
Definition at line 214 of file test_distance_table.cpp.
References ParticleSet::addTable(), CHECK(), doc, ParticleSet::getDistTableAB(), OhmmsElementBase::getName(), Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), ParticleSet::getTotalNum(), ParticleSet::GroupID, ParticleSet::isSameMass(), okay, Libxml2Document::parseFromString(), pow(), XMLParticleParser::readXML(), REQUIRE(), sqrt(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "TwoBodyJastrow other variables" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 215 of file test_J2_derivatives.cpp.
References TwoBodyJastrow< FT >::addFunc(), CHECK(), TwoBodyJastrow< FT >::checkOutVariables(), TwoBodyJastrow< FT >::evaluateDerivatives(), TwoBodyJastrow< FT >::evaluateDerivativesWF(), get_two_species_particleset(), TwoBodyJastrow< FT >::getComponentOffset(), VariableSet::insert(), VariableSet::insertFrom(), VariableSet::resetIndex(), and VariableSet::size_of_active().
| qmcplusplus::TEST_CASE | ( | "SOVMC-alle" | , |
| "" | [drivers][vmc] | ||
| ) |
Definition at line 215 of file test_vmc_driver.cpp.
References CloneManager::acceptRatio(), SpeciesSet::addAttribute(), TrialWaveFunction::addComponent(), QMCHamiltonian::addOperator(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), CloneManager::clearClones(), OHMMS::Controller, ParticleSet::create(), MCWalkerConfiguration::createWalkers(), doc, Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), ParticleSet::getSpeciesSet(), okay, omp_get_max_threads(), Libxml2Document::parseFromString(), QMCDriver::process(), ParticleSet::R, REQUIRE(), MCWalkerConfiguration::resetWalkerProperty(), VMC::run(), Communicate::setName(), ParticleSet::setName(), ParticleSet::setSpinor(), ParticleSet::spins, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "J1 evaluate derivatives Jastrow with two species one without Jastrow" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 216 of file test_J1OrbitalSoA.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), ParticleSet::addTable(), app_log(), WaveFunctionFactory::buildTWF(), CHECK(), OHMMS::Controller, ParticleSet::create(), doc, ParticleSet::get(), ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), ispecies, okay, Libxml2Document::parseFromString(), ParticleSet::R, VariableSet::removeInactive(), REQUIRE(), ParticleSet::resetGroups(), ParticleSet::setName(), VariableSet::size_of_active(), twf, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "RandomForTest_call_operator" | , |
| "" | [utilities][for_testing] | ||
| ) |
Definition at line 216 of file test_RandomForTest.cpp.
References CHECK().
| qmcplusplus::TEST_CASE | ( | "TrialWaveFunction flex_evaluateDeltaLogSetup" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 217 of file test_TrialWaveFunction_He.cpp.
References TrialWaveFunction::addComponent(), CHECK(), OHMMS::Controller, convertUPtrToRefVector(), create_particle_gradient(), create_particle_laplacian(), TrialWaveFunction::createResource(), ParticleSet::createResource(), TrialWaveFunction::evaluateDeltaLog(), TrialWaveFunction::evaluateDeltaLogSetup(), ParticleSet::G, TrialWaveFunction::getOrbitals(), ParticleSet::L, TrialWaveFunction::mw_evaluateDeltaLog(), TrialWaveFunction::mw_evaluateDeltaLogSetup(), ParticleSet::R, Vector< T, Alloc >::resize(), TrialWaveFunction::setLogPsi(), setup_He_wavefunction(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "MomentumDistribution::spawnCrowdClone" | , |
| "" | [estimators] | ||
| ) |
Definition at line 219 of file test_MomentumDistribution.cpp.
References ProjectData::BATCH, clone, comm, OHMMS::Controller, crowd, doc, Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), lattice, MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), qmcplusplus::testing::makeTestLattice(), node, okay, Libxml2Document::parseFromString(), particle_pool, pset, rank, REQUIRE(), test_project, and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "DiracMatrix_inverse_complex" | , |
| "" | [wavefunction][fermion] | ||
| ) |
This test case is meant to match the cuBLAS_LU::getrf_batched_complex.
Definition at line 220 of file test_DiracMatrix.cpp.
References CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), Matrix< T, Alloc >::cols(), Matrix< T, Alloc >::data(), DiracMatrix< T_FP >::invert_transpose(), lu, lu_mat(), Matrix< T, Alloc >::resize(), Matrix< T, Alloc >::rows(), qmcplusplus::simd::transpose(), and Xgetrf().
| qmcplusplus::TEST_CASE | ( | "Rotated LCAO WF0 zero angle" | , |
| "" | [qmcapp] | ||
| ) |
Definition at line 224 of file test_RotatedSPOs_LCAO.cpp.
References ProjectData::BATCH, CHECK(), TrialWaveFunction::checkInVariables(), TrialWaveFunction::checkOutVariables(), OHMMS::Controller, doc, TrialWaveFunction::evaluateDerivatives(), TrialWaveFunction::evaluateLog(), ParticleSet::G, TrialWaveFunction::getOrbitals(), ParticleSetPool::getParticleSet(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), identity_coeff, ParticleSet::L, TrialWaveFunction::mw_evaluateParameterDerivatives(), okay, Libxml2Document::parseFromString(), REQUIRE(), TrialWaveFunction::resetParameters(), setupParticleSetPool(), setupRotationXML(), test_project, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "test_timer_nested_profile_alt_routes" | , |
| "" | [utilities] | ||
| ) |
Definition at line 228 of file test_timer.cpp.
References CHECK(), TimerManager< TIMER >::collate_stack_profile(), convert_to_ns(), TimerManager< TIMER >::createTimer(), doc, Libxml2Document::dump(), Libxml2Document::getRoot(), TimerManager< TIMER >::StackProfileData::names, Libxml2Document::newDoc(), TimerManager< TIMER >::output_timing(), REQUIRE(), qmcplusplus::Units::time::s, TimerManager< TIMER >::set_timer_threshold(), TimerType< CLOCK >::start(), TimerType< CLOCK >::stop(), TimerManager< TIMER >::StackProfileData::timeExclList, TimerManager< TIMER >::StackProfileData::timeList, and timer_level_fine.
| qmcplusplus::TEST_CASE | ( | "benchmark_DiracMatrixComputeCUDASingle_vs_legacy_1024_4" | , |
| "" | [wavefunction][fermion][.benchmark] | ||
| ) |
Only runs if [benchmark] tag is passed.
Definition at line 230 of file benchmark_DiracMatrixComputeCUDA.cpp.
References DiracComputeBenchmarkParameters::batch_size, DiracMatrix< T_FP >::invert_transpose(), DiracMatrixComputeCUDA< VALUE_FP >::invert_transpose(), log_values(), qmcplusplus::testing::makeRngSpdMatrix(), qmcplusplus::Units::meter, DiracComputeBenchmarkParameters::n, DiracComputeBenchmarkParameters::name, queue, and DiracComputeBenchmarkParameters::str().
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A Ewald Quasi2D staggered triangle" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 231 of file test_ewald_quasi2d.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), dot(), CoulombPBCAA::evaluate(), ParticleSet::getSpeciesSet(), lattice, LRCoulombSingleton::QUASI2D, ParticleSet::R, ParticleSet::setName(), sqrt(), LRCoulombSingleton::this_lr_type, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "HCN MO with cusp" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 235 of file test_soa_cusp_corr.cpp.
References castCharToXMLChar(), CHECK(), OHMMS::Controller, SPOSetBuilderFactory::createSPOSetBuilder(), doc, Libxml2Document::getRoot(), Libxml2Document::getXPathContext(), okay, Libxml2Document::parse(), XMLParticleParser::readXML(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "OneBodyDensityMatrices::OneBodyDensityMatrices" | , |
| "" | [estimators] | ||
| ) |
Definition at line 238 of file test_OneBodyDensityMatrices.cpp.
References ProjectData::BATCH, comm, OHMMS::Controller, doc, Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), lattice, MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), qmcplusplus::testing::makeSpeciesSet(), qmcplusplus::testing::makeTestLattice(), node, obdmi, oeba, okay, Libxml2Document::parseFromString(), particle_pool, pset_target, species_set, test_project, OneBodyDensityMatricesTests< T >::testCopyConstructor(), and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "SPO input spline from xml LiH_msd arbitrary species" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 253 of file test_spo_collection_input_MSD_LCAO_h5.cpp.
References app_log(), and test_LiH_msd_xml_input_with_positron().
| qmcplusplus::TEST_CASE | ( | "PolynomialFunctor3D Jastrow" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 254 of file test_polynomial_eeI_jastrow.cpp.
References DC_POS, DC_POS_OFFLOAD, and test_J3_polynomial3D().
| qmcplusplus::TEST_CASE | ( | "Ceperley Force" | , |
| "" | [hamiltonian] | ||
| ) |
From the 'Force.ipynb' Jupyter notebook
Definition at line 255 of file test_force.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ForceCeperley::c, CHECK(), ParticleSet::create(), qmcplusplus::Units::charge::e, ForceCeperley::evaluate(), ForceBase::getForces(), ForceBase::getForcesIonIon(), ParticleSet::getSpeciesSet(), ForceCeperley::InitMatrix(), ForceCeperley::N_basis, ParticleSet::R, ForceCeperley::Rcut, ParticleSet::resetGroups(), ForceBase::setAddIonIon(), ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "Einspline SPO from HDF diamond_2x1x1 5 electrons" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 261 of file test_einset.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), CHECK(), OHMMS::Controller, ParticleSet::create(), ParticleSet::createResource(), EinsplineSetBuilder::createSPOSetFromXML(), Vector< T, Alloc >::device_data(), QMCTraits::DIM_VGL, doc, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), imag(), lattice, okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), Array< T, D, ALLOC >::resize(), ParticleSet::setName(), ParticleSetPool::setSimulationCell(), Vector< T, Alloc >::size(), and Vector< T, Alloc >::updateTo().
| qmcplusplus::TEST_CASE | ( | "DiracDeterminantBatched_second" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 263 of file test_DiracDeterminantBatched.cpp.
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A BCC 3 particles" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 267 of file test_coulomb_pbcAA.cpp.
References DC_POS, DC_POS_OFFLOAD, and test_CoulombPBCAA_3p().
| qmcplusplus::TEST_CASE | ( | "InputSection::InvalidElement" | , |
| "" | [estimators] | ||
| ) |
Definition at line 273 of file test_InputSection.cpp.
References doc, Libxml2Document::getRoot(), okay, Libxml2Document::parseFromString(), InputSection::readXML(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "OneBodyDensityMatrices::generateSamples" | , |
| "" | [estimators] | ||
| ) |
Definition at line 274 of file test_OneBodyDensityMatrices.cpp.
References ProjectData::BATCH, comm, OHMMS::Controller, doc, Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), node, obdmi, okay, Libxml2Document::parseFromString(), particle_pool, pset_target, species_set, test_project, and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "CoulombAA::mw_evaluatePerParticle" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 274 of file test_coulomb_pbcAA.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), Matrix< T, Alloc >::begin(), CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), Matrix< T, Alloc >::cols(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), CoulombPBCAA::createResource(), ParticleSet::createResource(), ParticleSet::createSK(), DC_POS, DC_POS_OFFLOAD, qmcplusplus::testing::getParticularListener(), ParticleSet::getSpeciesSet(), OperatorBase::getValue(), CoulombPBCAA::informOfPerParticleListener(), lattice, CoulombPBCAA::mw_evaluatePerParticle(), ParticleSet::mw_update(), CoulombPBCAA::myConst, ParticleSet::R, ParticleSet::setName(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "RotatedSPOs SplineR2R hcpBe values" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 274 of file test_RotatedSPOs_NLPP.cpp.
References test_hcpBe_rotation().
| qmcplusplus::TEST_CASE | ( | "test_timer_nested_profile_collate" | , |
| "" | [utilities] | ||
| ) |
Definition at line 280 of file test_timer.cpp.
References TimerManager< TIMER >::collate_stack_profile(), convert_to_ns(), TimerManager< TIMER >::createTimer(), doc, Libxml2Document::dump(), Libxml2Document::getRoot(), TimerManager< TIMER >::StackProfileData::names, Libxml2Document::newDoc(), TimerManager< TIMER >::output_timing(), REQUIRE(), qmcplusplus::Units::time::s, TimerManager< TIMER >::set_timer_threshold(), TimerType< CLOCK >::start(), TimerType< CLOCK >::stop(), and timer_level_fine.
| qmcplusplus::TEST_CASE | ( | "RotatedSPOs createRotationIndices" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 282 of file test_RotatedSPOs.cpp.
References CHECK(), RotatedSPOs::createRotationIndices(), and RotatedSPOs::createRotationIndicesFull().
| qmcplusplus::TEST_CASE | ( | "mw_evaluate Numerical He" | , |
| "" | [wavefunction] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "InputSection::init" | , |
| "" | [estimators] | ||
| ) |
Definition at line 284 of file test_InputSection.cpp.
References InputSection::init().
| qmcplusplus::TEST_CASE | ( | "DiracDeterminant_second" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 285 of file test_DiracDeterminant.cpp.
| qmcplusplus::TEST_CASE | ( | "SOECPotential" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 285 of file test_SOECPotential.cpp.
References doSOECPotentialTest().
| qmcplusplus::TEST_CASE | ( | "SPO input spline from h5 LiH_msd arbitrary species" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 285 of file test_spo_collection_input_MSD_LCAO_h5.cpp.
References app_log(), and test_LiH_msd_xml_input_with_positron().
| qmcplusplus::TEST_CASE | ( | "Cartesian Tensor evaluateWithThirdDeriv subset" | , |
| "" | [numerics] | ||
| ) |
Definition at line 287 of file test_cartesian_tensor.cpp.
References CHECK(), CartesianTensor< T, Point_t, Tensor_t, GGG_t >::evaluateWithThirdDeriv(), CartesianTensor< T, Point_t, Tensor_t, GGG_t >::getGGGYlm(), CartesianTensor< T, Point_t, Tensor_t, GGG_t >::getGradYlm(), CartesianTensor< T, Point_t, Tensor_t, GGG_t >::getHessYlm(), and CartesianTensor< T, Point_t, Tensor_t, GGG_t >::getYlm().
| qmcplusplus::TEST_CASE | ( | "Coulomb PBC A-A Ewald Quasi2D staggered triangle 2x2" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 287 of file test_ewald_quasi2d.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), ParticleSet::createSK(), dot(), CoulombPBCAA::evaluate(), for(), ParticleSet::getSpeciesSet(), lattice, LRCoulombSingleton::QUASI2D, ParticleSet::R, ParticleSet::setName(), sqrt(), LRCoulombSingleton::this_lr_type, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "TwoBodyJastrow Jastrow three particles of three types" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 288 of file test_J2_derivatives.cpp.
References TwoBodyJastrow< FT >::addFunc(), CHECK(), ParticleSet::create(), TwoBodyJastrow< FT >::getPairFunctions(), ParticleSet::R, and ParticleSet::setName().
| qmcplusplus::TEST_CASE | ( | "cuBLAS_LU::getrf_batched_complex" | , |
| "" | [wavefunction][CUDA] | ||
| ) |
Definition at line 295 of file test_cuBLAS_LU.cpp.
References qmcplusplus::Units::distance::A, B(), batch_size, CHECK(), check_matrix_result, checkArray, CHECKED_ELSE(), checkMatrix(), qmcplusplus::cuBLAS_LU::computeGetrf_batched(), cudaErrorCheck(), cudaMemcpyAsync, cudaMemcpyDeviceToHost, cudaMemcpyHostToDevice, cudaStreamSynchronize, dev_infos(), dev_pivots(), devMs(), hstream, infos(), lda, lu, lu_mat(), M_mat(), Ms, n, pivots, and real_pivot.
| qmcplusplus::TEST_CASE | ( | "Rotated LCAO WF1" | , |
| "" | [qmcapp] | ||
| ) |
Definition at line 298 of file test_RotatedSPOs_LCAO.cpp.
References ProjectData::BATCH, CHECK(), TrialWaveFunction::checkInVariables(), TrialWaveFunction::checkOutVariables(), OHMMS::Controller, doc, TrialWaveFunction::evaluateDerivatives(), TrialWaveFunction::evaluateLog(), ParticleSet::G, TrialWaveFunction::getOrbitals(), ParticleSetPool::getParticleSet(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), identity_coeff, ParticleSet::L, okay, Libxml2Document::parseFromString(), REQUIRE(), TrialWaveFunction::resetParameters(), setupParticleSetPool(), setupRotationXML(), test_project, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "SpaceGrid::Accumulate::outside" | , |
| "" | [estimators] | ||
| ) |
Definition at line 314 of file test_NESpaceGrid.cpp.
References ParticleSet::addTable(), ParticleSet::applyMinimumImage(), CHECK(), comm, OHMMS::Controller, ParticleSet::getDistTableAB(), ParticleSet::getTotalNum(), OHMMS_DIM, SpaceGridEnv< VALID >::pset_elec_, SpaceGridEnv< VALID >::pset_ions_, ParticleSet::R, SpaceGridEnv< VALID >::ref_points_, Matrix< T, Alloc >::resize(), SpaceGridEnv< VALID >::sgi_, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "DiracMatrix_update_row" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 315 of file test_DiracMatrix.cpp.
References DelayedUpdate< T, T_FP >::acceptRow(), CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), Matrix< T, Alloc >::cols(), Matrix< T, Alloc >::data(), Vector< T, Alloc >::data(), qmcplusplus::simd::dot(), DelayedUpdate< T, T_FP >::getInvRow(), DiracMatrix< T_FP >::invert_transpose(), DelayedUpdate< T, T_FP >::resize(), Matrix< T, Alloc >::resize(), Matrix< T, Alloc >::rows(), Vector< T, Alloc >::size(), and qmcplusplus::simd::transpose().
| qmcplusplus::TEST_CASE | ( | "RotatedSPOs constructAntiSymmetricMatrix" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 317 of file test_RotatedSPOs.cpp.
References CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), RotatedSPOs::constructAntiSymmetricMatrix(), RotatedSPOs::createRotationIndices(), and RotatedSPOs::extractParamsFromAntiSymmetricMatrix().
| qmcplusplus::TEST_CASE | ( | "Ion-ion Force" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 325 of file test_force.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), ParticleSet::create(), ForceBase::getForces(), ParticleSet::getSpeciesSet(), ParticleSet::R, ParticleSet::resetGroups(), and ParticleSet::setName().
| qmcplusplus::TEST_CASE | ( | "OmpBLAS ger" | , |
| "" | [SYCL] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "Eloc_Derivatives:slater_wj" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 332 of file test_ion_derivs.cpp.
References app_log(), WaveFunctionFactory::buildTWF(), CHECK(), OHMMS::Controller, convertToReal(), create_CN_Hamiltonian(), create_CN_particlesets(), dot(), qmcplusplus::Units::charge::e, TrialWaveFunction::evalGradSource(), QMCHamiltonian::evaluateDeterministic(), QMCHamiltonian::evaluateIonDerivs(), QMCHamiltonian::evaluateIonDerivsDeterministic(), TrialWaveFunction::evaluateLog(), QMCHamiltonian::getHamiltonian(), QMCHamiltonian::getObservable(), QMCHamiltonian::getObservableName(), Libxml2Document::getRoot(), ham, Libxml2Document::parse(), REQUIRE(), Vector< T, Alloc >::resize(), QMCHamiltonian::setRandomGenerator(), and QMCHamiltonian::sizeOfObservables().
| qmcplusplus::TEST_CASE | ( | "LiH multi Slater dets table_method" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 340 of file test_multi_slater_determinant.cpp.
References app_log(), and test_LiH_msd().
| qmcplusplus::TEST_CASE | ( | "ReferencePoints::HDF5" | , |
| "" | [estimators] | ||
| ) |
Definition at line 343 of file test_ReferencePoints.cpp.
References approxEquality(), CHECK(), hdf_archive::close(), hdf_archive::create(), expectedReferencePoints(), makePsets(), makeTestRPI(), okay, hdf_archive::open(), hdf_archive::push(), hdf_archive::readEntry(), REQUIRE(), and NEReferencePoints::write().
| qmcplusplus::TEST_CASE | ( | "ompBLAS gemv" | , |
| "" | [OMP] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "BareKineticEnergyListener" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 351 of file test_bare_kinetic.cpp.
References getTestCaseForWeights(), outputManager, OutputManagerClass::pause(), OutputManagerClass::resume(), and testElecCase().
| qmcplusplus::TEST_CASE | ( | "RotatedSPOs exponentiate matrix" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 352 of file test_RotatedSPOs.cpp.
References CHECK(), CHECKED_ELSE(), checkMatrix(), RotatedSPOs::exponentiate_antisym_matrix(), CheckMatrixResult::result, and CheckMatrixResult::result_message.
| qmcplusplus::TEST_CASE | ( | "Rotated LCAO WF2 with jastrow" | , |
| "" | [qmcapp] | ||
| ) |
Definition at line 355 of file test_RotatedSPOs_LCAO.cpp.
References ProjectData::BATCH, CHECK(), TrialWaveFunction::checkInVariables(), TrialWaveFunction::checkOutVariables(), OHMMS::Controller, doc, TrialWaveFunction::evaluateDerivatives(), TrialWaveFunction::evaluateLog(), ParticleSet::G, TrialWaveFunction::getOrbitals(), ParticleSetPool::getParticleSet(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), ParticleSet::L, okay, Libxml2Document::parseFromString(), REQUIRE(), TrialWaveFunction::resetParameters(), setupParticleSetPool(), test_project, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "SpaceGrid::BadPeriodic" | , |
| "" | [estimators] | ||
| ) |
Definition at line 365 of file test_NESpaceGrid.cpp.
References ParticleSet::addTable(), CHECK(), comm, OHMMS::Controller, ParticleSet::getDistTableAB(), ParticleSet::getTotalNum(), OHMMS_DIM, SpaceGridEnv< VALID >::pset_elec_, SpaceGridEnv< VALID >::pset_ions_, ParticleSet::R, SpaceGridEnv< VALID >::ref_points_, Matrix< T, Alloc >::resize(), SpaceGridEnv< VALID >::sgi_, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "InputSection::get" | , |
| "" | [estimators] | ||
| ) |
Definition at line 366 of file test_InputSection.cpp.
References InputSection::get(), and InputSection::init().
| qmcplusplus::TEST_CASE | ( | "ompBLAS gemv notrans" | , |
| "" | [OMP] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "LiH multi Slater dets precomputed_table_method" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 369 of file test_multi_slater_determinant.cpp.
References app_log(), and test_LiH_msd().
| qmcplusplus::TEST_CASE | ( | "Spline applyRotation two rotations" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 375 of file test_spline_applyrotation.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), CHECK(), CHECKED_ELSE(), checkMatrix(), OHMMS::Controller, ParticleSet::create(), EinsplineSetBuilder::createSPOSetFromXML(), doc, qmcplusplus::Units::charge::e, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), lattice, norm(), okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), ParticleSet::setName(), ParticleSetPool::setSimulationCell(), Vector< T, Alloc >::size(), and sqrt().
| qmcplusplus::TEST_CASE | ( | "AC Force" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 376 of file test_force.cpp.
References ACForce::add2Hamiltonian(), SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), CHECK(), clone, ACForce::compute_regularizer_f(), ParticleSet::create(), ACForce::evaluate(), ACForce::get(), Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), ParticleSet::getTotalNum(), ACForce::makeClone(), Libxml2Document::parseFromString(), ACForce::put(), ParticleSet::R, REQUIRE(), ParticleSet::resetGroups(), ACForce::resetTargetParticleSet(), ParticleSet::setName(), sqrt(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "OneBodyDensityMatrices::accumulate" | , |
| "" | [estimators] | ||
| ) |
Definition at line 382 of file test_OneBodyDensityMatrices.cpp.
References ProjectData::BATCH, comm, OHMMS::Controller, convertUPtrToRefVector(), doc, dump_obdm, OneBodyDensityMatricesTests< T >::dumpData(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), StdRandom< T >::init(), MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), node, obdmi, okay, Libxml2Document::parseFromString(), particle_pool, pset_target, species_set, spomap, test_project, OneBodyDensityMatricesTests< T >::testAccumulate(), walker, qmcplusplus::hdf::walkers, and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "Cartesian Tensor evaluateThirdDerivOnly subset" | , |
| "" | [numerics] | ||
| ) |
Definition at line 391 of file test_cartesian_tensor.cpp.
References CHECK(), CartesianTensor< T, Point_t, Tensor_t, GGG_t >::evaluateThirdDerivOnly(), and CartesianTensor< T, Point_t, Tensor_t, GGG_t >::getGGGYlm().
| qmcplusplus::TEST_CASE | ( | "test setup timers" | , |
| "" | [utilities] | ||
| ) |
Definition at line 399 of file test_timer.cpp.
References CHECK(), convert_to_ns(), MyTimer1, REQUIRE(), qmcplusplus::Units::time::s, TestTimerNames, and timer_level_coarse.
| qmcplusplus::TEST_CASE | ( | "SpaceGrid::hdf5" | , |
| "" | [estimators] | ||
| ) |
Definition at line 405 of file test_NESpaceGrid.cpp.
References ParticleSet::addTable(), CHECK(), hdf_archive::close(), comm, OHMMS::Controller, hdf_archive::create(), ParticleSet::getDistTableAB(), ParticleSet::getTotalNum(), OHMMS_DIM, okay, hdf_archive::open(), SpaceGridEnv< VALID >::pset_elec_, SpaceGridEnv< VALID >::pset_ions_, hdf_archive::push(), ParticleSet::R, hdf_archive::readEntry(), SpaceGridEnv< VALID >::ref_points_, REQUIRE(), Matrix< T, Alloc >::resize(), SpaceGridEnv< VALID >::sgi_, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "Ethanol MO with cusp" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 410 of file test_soa_cusp_corr.cpp.
References castCharToXMLChar(), CHECK(), OHMMS::Controller, SPOSetBuilderFactory::createSPOSetBuilder(), doc, Libxml2Document::getRoot(), Libxml2Document::getXPathContext(), okay, Libxml2Document::parse(), XMLParticleParser::readXML(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "RotatedSPOs log matrix" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 424 of file test_RotatedSPOs.cpp.
References CHECK(), CHECKED_ELSE(), checkMatrix(), RotatedSPOs::log_antisym_matrix(), CheckMatrixResult::result, and CheckMatrixResult::result_message.
| qmcplusplus::TEST_CASE | ( | "AccelBLAS" | , |
| "" | [BLAS] | ||
| ) |
Definition at line 434 of file test_AccelBLAS.cpp.
| qmcplusplus::TEST_CASE | ( | "BSpline builder Jastrow J1" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 437 of file test_J1_bspline.cpp.
References DC_POS, DC_POS_OFFLOAD, and test_J1_spline().
| qmcplusplus::TEST_CASE | ( | "distance_pbc_z" | , |
| "" | [distance_table][xml] | ||
| ) |
Definition at line 444 of file test_distance_table.cpp.
References ParticleSet::accept_rejectMove(), ParticleSet::acceptMove(), ParticleSet::addTable(), CHECK(), ParticleSet::getDistTableAA(), ParticleSet::getDistTableAB(), ParticleSet::getTotalNum(), ParticleSet::makeMove(), parse_electron_ion_pbc_z(), parse_pbc_lattice(), ParticleSet::rejectMove(), and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "DiracDeterminant_delayed_update" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 444 of file test_DiracDeterminant.cpp.
| qmcplusplus::TEST_CASE | ( | "EinsplineSetBuilder CheckLattice" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 458 of file test_einset.cpp.
References EinsplineSetBuilder::CheckLattice(), OHMMS::Controller, lattice, REQUIRE(), and EinsplineSetBuilder::SuperLattice.
| qmcplusplus::TEST_CASE | ( | "cuBLAS_LU::getri_batched" | , |
| "" | [wavefunction][CUDA] | ||
| ) |
Definition at line 460 of file test_cuBLAS_LU.cpp.
References qmcplusplus::Units::distance::A, B(), batch_size, CHECK(), check_matrix_result, checkArray, CHECKED_ELSE(), checkMatrix(), qmcplusplus::cuBLAS_LU::computeGetri_batched(), cudaErrorCheck(), cudaMemcpyAsync, cudaMemcpyDeviceToHost, cudaMemcpyHostToDevice, cudaStreamSynchronize, dev_infos(), dev_pivots(), devM_vec(), devMs(), hstream, infos(), lda, M_vec, Ms, n, and pivots.
| qmcplusplus::TEST_CASE | ( | "mw_evaluate Numerical EtOH" | , |
| "" | [wavefunction] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "mw_evaluate GTO EtOH" | , |
| "" | [wavefunction] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "InputSection::custom" | , |
| "" | [estimators] | ||
| ) |
Definition at line 470 of file test_InputSection.cpp.
References CHECK(), doc, InputSection::get(), Libxml2Document::getRoot(), okay, Libxml2Document::parseFromString(), InputSection::readXML(), and CustomTestInput::report().
| qmcplusplus::TEST_CASE | ( | "OneBodyDensityMatrices::evaluateMatrix" | , |
| "" | [estimators] | ||
| ) |
Definition at line 472 of file test_OneBodyDensityMatrices.cpp.
References ProjectData::BATCH, comm, OHMMS::Controller, doc, dump_obdm, OneBodyDensityMatricesTests< T >::dumpData(), OneBodyDensityMatricesInput::get_integrator(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), StdRandom< T >::init(), OneBodyDensityMatricesInput::lookup_input_enum_value, MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), node, obdmi, okay, Libxml2Document::parseFromString(), particle_pool, pset_target, InputSection::reverseLookupInputEnumMap(), species_set, spomap, test_project, OneBodyDensityMatricesTests< T >::testEvaluateMatrix(), walker, and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "Rotated LCAO | WF1, |
| MO coeff | rotated, | ||
| zero angle" | , | ||
| "" | [qmcapp] | ||
| ) |
Definition at line 475 of file test_RotatedSPOs_LCAO.cpp.
References ProjectData::BATCH, CHECK(), TrialWaveFunction::checkInVariables(), TrialWaveFunction::checkOutVariables(), coeff_rot_by_point1, coeff_rot_by_point2, OHMMS::Controller, doc, TrialWaveFunction::evaluateDerivatives(), TrialWaveFunction::evaluateLog(), ParticleSet::G, TrialWaveFunction::getOrbitals(), ParticleSetPool::getParticleSet(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), ParticleSet::L, okay, Libxml2Document::parseFromString(), REQUIRE(), TrialWaveFunction::resetParameters(), setupParticleSetPool(), setupRotationXML(), test_project, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "DiracDeterminantBatched_delayed_update" | , |
| "" | [wavefunction][fermion] | ||
| ) |
Definition at line 477 of file test_DiracDeterminantBatched.cpp.
Definition at line 497 of file test_RotatedSPOs.cpp.
References CHECK(), CHECKED_ELSE(), checkMatrix(), RotatedSPOs::constructAntiSymmetricMatrix(), RotatedSPOs::createRotationIndices(), RotatedSPOs::exponentiate_antisym_matrix(), RotatedSPOs::extractParamsFromAntiSymmetricMatrix(), RotatedSPOs::log_antisym_matrix(), CheckMatrixResult::result, and CheckMatrixResult::result_message.
| qmcplusplus::TEST_CASE | ( | "Eloc_Derivatives:multislater_noj" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 500 of file test_ion_derivs.cpp.
References app_log(), WaveFunctionFactory::buildTWF(), CHECK(), OHMMS::Controller, convertToReal(), create_CN_Hamiltonian(), create_CN_particlesets(), qmcplusplus::Units::charge::e, TrialWaveFunction::evalGradSource(), QMCHamiltonian::evaluateDeterministic(), TrialWaveFunction::evaluateLog(), QMCHamiltonian::getObservable(), QMCHamiltonian::getObservableName(), Libxml2Document::getRoot(), ham, Libxml2Document::parse(), REQUIRE(), Vector< T, Alloc >::resize(), and QMCHamiltonian::sizeOfObservables().
| qmcplusplus::TEST_CASE | ( | "OneBodyDensityMatrices::registerAndWrite" | , |
| "" | [estimators] | ||
| ) |
Definition at line 527 of file test_OneBodyDensityMatrices.cpp.
References ProjectData::BATCH, comm, OHMMS::Controller, doc, OneBodyDensityMatricesInput::get_integrator(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), OneBodyDensityMatricesInput::lookup_input_enum_value, MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_diamondC_1x1x1(), node, obdmi, okay, Libxml2Document::parseFromString(), particle_pool, pset_target, InputSection::reverseLookupInputEnumMap(), species_set, spomap, test_project, OneBodyDensityMatricesTests< T >::testRegisterAndWrite(), and wavefunction_pool.
| qmcplusplus::TEST_CASE | ( | "ompBLAS ger" | , |
| "" | [OMP] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "Rotated LCAO WF1 MO coeff | rotated, |
| half angle" | , | ||
| "" | [qmcapp] | ||
| ) |
Definition at line 539 of file test_RotatedSPOs_LCAO.cpp.
References ProjectData::BATCH, CHECK(), TrialWaveFunction::checkInVariables(), TrialWaveFunction::checkOutVariables(), coeff_rot_by_point05, coeff_rot_by_point1, OHMMS::Controller, doc, TrialWaveFunction::evaluateDerivatives(), TrialWaveFunction::evaluateLog(), ParticleSet::G, TrialWaveFunction::getOrbitals(), ParticleSetPool::getParticleSet(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), ParticleSet::L, okay, Libxml2Document::parseFromString(), REQUIRE(), TrialWaveFunction::resetParameters(), setupParticleSetPool(), setupRotationXML(), test_project, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "broadcastCuspInfo" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 551 of file test_soa_cusp_corr.cpp.
References CuspCorrectionParameters::alpha, broadcastCuspInfo(), CuspCorrectionParameters::C, CHECK(), OHMMS::Controller, Communicate::rank(), CuspCorrectionParameters::Rc, CuspCorrectionParameters::redo, REQUIRE(), and CuspCorrectionParameters::sg.
| qmcplusplus::TEST_CASE | ( | "RotatedSPOs hcpBe" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 561 of file test_RotatedSPOs.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), CHECK(), OHMMS::Controller, ParticleSet::create(), EinsplineSetBuilder::createSPOSetFromXML(), doc, qmcplusplus::Units::charge::e, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), lattice, okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), VariableSet::resetIndex(), ParticleSet::setName(), ParticleSetPool::setSimulationCell(), and Vector< T, Alloc >::size().
| qmcplusplus::TEST_CASE | ( | "TrialWaveFunction_diamondC_2x1x1" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 564 of file test_TrialWaveFunction_diamondC_2x1x1.cpp.
| qmcplusplus::TEST_CASE | ( | "ReadMolecularOrbital GTO Ne" | , |
| "" | [wavefunction] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "ReadMolecularOrbital Numerical Ne" | , |
| "" | [wavefunction] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "ReadMolecularOrbital HCN" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 593 of file test_MO.cpp.
| qmcplusplus::TEST_CASE | ( | "InputSection::Delegate" | , |
| "" | [estimators] | ||
| ) |
Definition at line 595 of file test_InputSection.cpp.
References CHECK(), doc, DelegatingInput::get(), Libxml2Document::getRoot(), DelegatingInput::has(), okay, Libxml2Document::parseFromString(), and REQUIRE().
| qmcplusplus::TEST_CASE | ( | "Rotated LCAO rotation consistency" | , |
| "" | [qmcapp] | ||
| ) |
Definition at line 596 of file test_RotatedSPOs_LCAO.cpp.
References RotatedSPOs::apply_rotation(), RotatedSPOs::applyDeltaRotation(), ProjectData::BATCH, LCAOrbitalSet::C, CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), OHMMS::Controller, doc, TrialWaveFunction::getOrbitals(), SlaterDet::getPhi(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), okay, Libxml2Document::parseFromString(), RotatedSPOs::Phi_, REQUIRE(), CheckMatrixResult::result, CheckMatrixResult::result_message, setupParticleSetPool(), and test_project.
| qmcplusplus::TEST_CASE | ( | "Eloc_Derivatives:multislater_wj" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 639 of file test_ion_derivs.cpp.
References app_log(), WaveFunctionFactory::buildTWF(), CHECK(), OHMMS::Controller, convertToReal(), create_CN_Hamiltonian(), create_CN_particlesets(), qmcplusplus::Units::charge::e, TrialWaveFunction::evalGradSource(), QMCHamiltonian::evaluateDeterministic(), TrialWaveFunction::evaluateLog(), QMCHamiltonian::getObservable(), QMCHamiltonian::getObservableName(), Libxml2Document::getRoot(), ham, Libxml2Document::parse(), REQUIRE(), Vector< T, Alloc >::resize(), and QMCHamiltonian::sizeOfObservables().
| qmcplusplus::TEST_CASE | ( | "distance_pbc_z batched APIs" | , |
| "" | [distance_table][xml] | ||
| ) |
Definition at line 658 of file test_distance_table.cpp.
References DC_POS, DC_POS_OFFLOAD, test_distance_fcc_pbc_z_batched_APIs(), and test_distance_pbc_z_batched_APIs().
| qmcplusplus::TEST_CASE | ( | "RotatedSPOs construct delta matrix" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 674 of file test_RotatedSPOs.cpp.
References CHECK(), CHECKED_ELSE(), checkMatrix(), RotatedSPOs::constructDeltaRotation(), RotatedSPOs::createRotationIndices(), and RotatedSPOs::createRotationIndicesFull().
| qmcplusplus::TEST_CASE | ( | "distance_pbc_z batched APIs ee NEED_TEMP_DATA_ON_HOST" | , |
| "" | [distance_table][xml] | ||
| ) |
Definition at line 728 of file test_distance_table.cpp.
References DC_POS, DC_POS_OFFLOAD, and test_distance_pbc_z_batched_APIs_ee_NEED_TEMP_DATA_ON_HOST().
| qmcplusplus::TEST_CASE | ( | "test_distance_pbc_diamond" | , |
| "" | [distance_table][xml] | ||
| ) |
Definition at line 734 of file test_distance_table.cpp.
References OHMMS::Controller, and MinimalParticlePool::make_diamondC_1x1x1().
| qmcplusplus::TEST_CASE | ( | "RotatedSPOs read and write parameters" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 738 of file test_RotatedSPOs.cpp.
References RotatedSPOs::buildOptVariables(), CHECK(), RotatedSPOs::checkInVariablesExclusive(), qmcplusplus::testing::getMyVars(), qmcplusplus::testing::getMyVarsFull(), VariableSet::readFromHDF(), RotatedSPOs::readVariationalParameters(), RotatedSPOs::resetParametersExclusive(), VariableSet::size(), VariableSet::writeToHDF(), and RotatedSPOs::writeVariationalParameters().
| qmcplusplus::TEST_CASE | ( | "Rotated LCAO Be single determinant" | , |
| "" | [qmcapp] | ||
| ) |
Definition at line 745 of file test_RotatedSPOs_LCAO.cpp.
References ProjectData::BATCH, CHECK(), TrialWaveFunction::checkInVariables(), TrialWaveFunction::checkOutVariables(), OHMMS::Controller, doc, TrialWaveFunction::evaluateDerivatives(), TrialWaveFunction::evaluateLog(), TrialWaveFunction::getOrbitals(), ParticleSetPool::getParticleSet(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), okay, Libxml2Document::parse(), REQUIRE(), TrialWaveFunction::resetParameters(), setupParticleSetPoolBe(), test_project, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "ReadMolecularOrbital GTO spinor" | , |
| "" | [wavefunction] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "ReadMolecularOrbital GTO spinor with excited" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 765 of file test_MO_spinor.cpp.
References test_lcao_spinor_excited().
| qmcplusplus::TEST_CASE | ( | "spinor ion derivatives for molecule" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 766 of file test_MO_spinor.cpp.
References test_lcao_spinor_ion_derivs().
| qmcplusplus::TEST_CASE | ( | "Eloc_Derivatives:proto_sd_noj" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 778 of file test_ion_derivs.cpp.
References app_log(), B(), WaveFunctionFactory::buildTWF(), CHECK(), OHMMS::Controller, convertToReal(), create_CN_Hamiltonian(), create_CN_particlesets(), qmcplusplus::Units::charge::e, OperatorBase::evaluateOneBodyOpMatrix(), OperatorBase::evaluateOneBodyOpMatrixForceDeriv(), QMCHamiltonian::getHamiltonian(), Libxml2Document::getRoot(), ham, TrialWaveFunction::initializeTWFFastDerivWrapper(), OHMMS_DIM, okay, Libxml2Document::parse(), REQUIRE(), and twf.
| qmcplusplus::TEST_CASE | ( | "RotatedSPOs read and write parameters history" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 790 of file test_RotatedSPOs.cpp.
References RotatedSPOs::buildOptVariables(), CHECK(), RotatedSPOs::checkInVariablesExclusive(), qmcplusplus::testing::getHistoryParams(), qmcplusplus::testing::getMyVars(), VariableSet::readFromHDF(), RotatedSPOs::readVariationalParameters(), REQUIRE(), RotatedSPOs::resetParametersExclusive(), RotatedSPOs::set_use_global_rotation(), VariableSet::size(), VariableSet::writeToHDF(), and RotatedSPOs::writeVariationalParameters().
| qmcplusplus::TEST_CASE | ( | "Rotated LCAO Be multi determinant with one determinant" | , |
| "" | [qmcapp] | ||
| ) |
Definition at line 813 of file test_RotatedSPOs_LCAO.cpp.
References ProjectData::BATCH, CHECK(), TrialWaveFunction::checkInVariables(), TrialWaveFunction::checkOutVariables(), OHMMS::Controller, doc, TrialWaveFunction::evaluateDerivatives(), TrialWaveFunction::evaluateLog(), TrialWaveFunction::getOrbitals(), ParticleSetPool::getParticleSet(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), okay, Libxml2Document::parse(), REQUIRE(), TrialWaveFunction::resetParameters(), setupParticleSetPoolBe(), test_project, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "ReadMolecularOrbital GTO Carbon Diamond" | , |
| "" | [wavefunction] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "Rotated LCAO Be two determinant" | , |
| "" | [qmcapp] | ||
| ) |
Definition at line 880 of file test_RotatedSPOs_LCAO.cpp.
References ProjectData::BATCH, CHECK(), TrialWaveFunction::checkInVariables(), TrialWaveFunction::checkOutVariables(), OHMMS::Controller, doc, TrialWaveFunction::evaluateDerivatives(), TrialWaveFunction::evaluateLog(), TrialWaveFunction::getOrbitals(), ParticleSetPool::getParticleSet(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), okay, Libxml2Document::parse(), REQUIRE(), TrialWaveFunction::resetParameters(), setupParticleSetPoolBe(), test_project, and ParticleSet::update().
| qmcplusplus::TEST_CASE | ( | "LCAOrbitalSet batched PBC DiamondC" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 913 of file test_LCAO_diamondC_2x1x1.cpp.
References test_LCAO_DiamondC_2x1x1_cplx(), and test_LCAO_DiamondC_2x1x1_real().
| qmcplusplus::TEST_CASE | ( | "LCAOrbitalSet batched PBC DiamondC offload" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 921 of file test_LCAO_diamondC_2x1x1.cpp.
References test_LCAO_DiamondC_2x1x1_cplx(), and test_LCAO_DiamondC_2x1x1_real().
| qmcplusplus::TEST_CASE | ( | "RotatedSPOs mw_ APIs" | , |
| "" | [wavefunction] | ||
| ) |
Definition at line 941 of file test_RotatedSPOs.cpp.
References CHECK(), RotatedSPOs::createResource(), RotatedSPOs::evaluate_spin(), qmcplusplus::Units::distance::m, RotatedSPOs::mw_evaluateValue(), and RotatedSPOs::mw_evaluateVGLWithSpin().
| qmcplusplus::TEST_CASE | ( | "ReadMolecularOrbital GTO HCN" | , |
| "" | [wavefunction] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "ReadMolecularOrbital Numerical HCN" | , |
| "" | [wavefunction] | ||
| ) |
| qmcplusplus::TEST_CASE | ( | "Eloc_Derivatives:proto_sd_wj" | , |
| "" | [hamiltonian] | ||
| ) |
Definition at line 1038 of file test_ion_derivs.cpp.
References app_log(), B(), WaveFunctionFactory::buildTWF(), CHECK(), OHMMS::Controller, convertToReal(), create_CN_Hamiltonian(), create_CN_particlesets(), qmcplusplus::Units::charge::e, QMCHamiltonian::evaluateIonDerivsDeterministicFast(), OperatorBase::evaluateOneBodyOpMatrix(), OperatorBase::evaluateOneBodyOpMatrixForceDeriv(), QMCHamiltonian::getHamiltonian(), Libxml2Document::getRoot(), ham, TrialWaveFunction::initializeTWFFastDerivWrapper(), OHMMS_DIM, okay, Libxml2Document::parse(), REQUIRE(), and twf.
| void qmcplusplus::test_CoulombPBCAA_3p | ( | DynamicCoordinateKind | kind | ) |
Definition at line 196 of file test_coulomb_pbcAA.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), LRCoulombSingleton::CoulombHandler, ParticleSet::create(), CoulombPBCAA::createResource(), ParticleSet::createResource(), ParticleSet::createSK(), DC_POS_OFFLOAD, CoulombPBCAA::get_madelung_constant(), ParticleSet::getSpeciesSet(), OperatorBase::getValue(), lattice, CoulombPBCAA::mw_evaluate(), ParticleSet::mw_update(), ParticleSet::R, and ParticleSet::setName().
Referenced by TEST_CASE().
| void qmcplusplus::test_diamond_2x1x1_xml_input | ( | const std::string & | spo_xml_string | ) |
Definition at line 30 of file test_spo_collection_input_spline.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), CHECK(), OHMMS::Controller, ParticleSet::create(), doc, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), imag(), lattice, okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), ParticleSet::setName(), ParticleSetPool::setSimulationCell(), and Vector< T, Alloc >::size().
Referenced by TEST_CASE().
| void qmcplusplus::test_dirac_ao | ( | ) |
Definition at line 94 of file test_cartesian_ao.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), CHECK(), OHMMS::Controller, SPOSetBuilderFactory::createSPOSetBuilder(), doc, Libxml2Document::getRoot(), Libxml2Document::getXPathContext(), ispecies, okay, Libxml2Document::parse(), and REQUIRE().
Referenced by TEST_CASE().
| void qmcplusplus::test_DiracDeterminant_delayed_update | ( | const DetMatInvertor | inverter_kind | ) |
Definition at line 301 of file test_DiracDeterminant.cpp.
References app_log(), CHECK(), check_matrix(), Matrix< T, Alloc >::cols(), LogToValue< T >::convert(), ParticleSet::create(), Matrix< T, Alloc >::data(), exp(), DiracMatrix< T_FP >::invert_transpose(), Matrix< T, Alloc >::resize(), Matrix< T, Alloc >::rows(), and qmcplusplus::simd::transpose().
| void qmcplusplus::test_DiracDeterminant_first | ( | const DetMatInvertor | inverter_kind | ) |
Definition at line 52 of file test_DiracDeterminant.cpp.
References ParticleSet::acceptMove(), CHECK(), check_matrix(), ParticleSet::create(), ParticleSet::getTotalNum(), ParticleSet::makeMove(), VirtualParticleSet::makeMoves(), ParticleSet::makeVirtualMoves(), ParticleSet::R, ParticleSet::rejectMove(), and Matrix< T, Alloc >::resize().
| void qmcplusplus::test_DiracDeterminant_second | ( | const DetMatInvertor | inverter_kind | ) |
Definition at line 160 of file test_DiracDeterminant.cpp.
References app_log(), CHECK(), check_matrix(), Matrix< T, Alloc >::cols(), LogToValue< T >::convert(), ParticleSet::create(), Matrix< T, Alloc >::data(), exp(), DiracMatrix< T_FP >::invert_transpose(), Matrix< T, Alloc >::resize(), Matrix< T, Alloc >::rows(), and qmcplusplus::simd::transpose().
| void qmcplusplus::test_DiracDeterminantBatched_delayed_update | ( | int | delay_rank, |
| DetMatInvertor | matrix_inverter_kind | ||
| ) |
Definition at line 275 of file test_DiracDeterminantBatched.cpp.
References app_log(), CHECK(), CHECKED_ELSE(), checkMatrix(), Matrix< T, Alloc >::cols(), LogToValue< T >::convert(), ParticleSet::create(), ParticleSet::createResource(), Matrix< T, Alloc >::data(), exp(), DiracMatrix< T_FP >::invert_transpose(), ParticleSet::mw_update(), Matrix< T, Alloc >::resize(), Matrix< T, Alloc >::rows(), and qmcplusplus::simd::transpose().
| void qmcplusplus::test_DiracDeterminantBatched_first | ( | ) |
Definition at line 37 of file test_DiracDeterminantBatched.cpp.
References ParticleSet::acceptMove(), CHECK(), CHECKED_ELSE(), checkMatrix(), ParticleSet::create(), ParticleSet::getTotalNum(), ParticleSet::makeMove(), VirtualParticleSet::makeMoves(), ParticleSet::makeVirtualMoves(), ParticleSet::R, ParticleSet::rejectMove(), and Matrix< T, Alloc >::resize().
| void qmcplusplus::test_DiracDeterminantBatched_second | ( | ) |
Definition at line 140 of file test_DiracDeterminantBatched.cpp.
References app_log(), CHECK(), CHECKED_ELSE(), checkMatrix(), Matrix< T, Alloc >::cols(), LogToValue< T >::convert(), ParticleSet::create(), Matrix< T, Alloc >::data(), exp(), DiracMatrix< T_FP >::invert_transpose(), Matrix< T, Alloc >::resize(), Matrix< T, Alloc >::rows(), and qmcplusplus::simd::transpose().
| void qmcplusplus::test_distance_fcc_pbc_z_batched_APIs | ( | DynamicCoordinateKind | test_kind | ) |
Definition at line 612 of file test_distance_table.cpp.
References ParticleSet::accept_rejectMove(), ParticleSet::addTable(), CHECK(), ParticleSet::createResource(), ParticleSet::donePbyP(), ParticleSet::getDistTableAA(), ParticleSet::makeMove(), ParticleSet::mw_accept_rejectMove(), ParticleSet::mw_donePbyP(), ParticleSet::mw_makeMove(), NEED_FULL_TABLE_ON_HOST_AFTER_DONEPBYP, parse_electron_ion_pbc_z(), parse_pbc_fcc_lattice(), and ParticleSet::update().
Referenced by TEST_CASE().
| void qmcplusplus::test_distance_pbc_z_batched_APIs | ( | DynamicCoordinateKind | test_kind | ) |
Definition at line 566 of file test_distance_table.cpp.
References ParticleSet::accept_rejectMove(), ParticleSet::addTable(), CHECK(), ParticleSet::createResource(), ParticleSet::donePbyP(), ParticleSet::getDistTableAA(), ParticleSet::makeMove(), ParticleSet::mw_accept_rejectMove(), ParticleSet::mw_donePbyP(), ParticleSet::mw_makeMove(), NEED_FULL_TABLE_ON_HOST_AFTER_DONEPBYP, parse_electron_ion_pbc_z(), parse_pbc_lattice(), and ParticleSet::update().
Referenced by TEST_CASE().
| void qmcplusplus::test_distance_pbc_z_batched_APIs_ee_NEED_TEMP_DATA_ON_HOST | ( | DynamicCoordinateKind | test_kind | ) |
Definition at line 666 of file test_distance_table.cpp.
References ParticleSet::accept_rejectMove(), ParticleSet::addTable(), CHECK(), ParticleSet::createResource(), ParticleSet::donePbyP(), ParticleSet::getDistTableAA(), ParticleSet::makeMove(), ParticleSet::mw_accept_rejectMove(), ParticleSet::mw_makeMove(), NEED_TEMP_DATA_ON_HOST, parse_electron_ion_pbc_z(), parse_pbc_lattice(), and ParticleSet::update().
Referenced by TEST_CASE().
| void qmcplusplus::test_e2iphi | ( | ) |
Definition at line 24 of file test_e2iphi.cpp.
References CHECK(), cos(), eval_e2iphi(), qmcplusplus::Units::force::N, and sin().
| void qmcplusplus::test_EtOH_mw | ( | bool | transform | ) |
Definition at line 285 of file test_MO.cpp.
References castCharToXMLChar(), CHECK(), OHMMS::Controller, SPOSetBuilderFactory::createSPOSetBuilder(), QMCTraits::DIM, doc, qmcplusplus::Units::charge::e, Libxml2Document::getRoot(), Libxml2Document::getXPathContext(), okay, Libxml2Document::parse(), ParticleSet::R, XMLParticleParser::readXML(), REQUIRE(), and ParticleSet::update().
Referenced by TEST_CASE().
| void qmcplusplus::test_gemm | ( | const int | M, |
| const int | N, | ||
| const int | K, | ||
| const char | transa, | ||
| const char | transb | ||
| ) |
Definition at line 26 of file test_ompBLAS.cpp.
References qmcplusplus::Units::distance::A, B(), qmcplusplus::Units::charge::C, CHECK(), Vector< T, Alloc >::device_data(), qmcplusplus::ompBLAS::gemm(), BLAS::gemm(), qmcplusplus::ompBLAS::gemm_batched(), imag(), qmcplusplus::Units::energy::K, qmcplusplus::Units::force::N, and Vector< T, Alloc >::updateTo().
| void qmcplusplus::test_gemm_cases | ( | ) |
Definition at line 139 of file test_AccelBLAS.cpp.
References qmcplusplus::Units::energy::K, and qmcplusplus::Units::force::N.
| void test_gemv | ( | const int | M_b, |
| const int | N_b, | ||
| const char | trans | ||
| ) |
Definition at line 184 of file test_ompBLAS.cpp.
References qmcplusplus::Units::distance::A, B(), qmcplusplus::Units::charge::C, CHECK(), TinyVector< T, D >::data(), qmcplusplus::ompBLAS::gemv(), BLAS::gemv(), BLAS::gemv_trans(), and qmcplusplus::Units::force::N.
| void test_gemv_batched | ( | const int | M_b, |
| const int | N_b, | ||
| const char | trans, | ||
| const int | batch_count | ||
| ) |
Definition at line 234 of file test_ompBLAS.cpp.
References CHECK(), Vector< T, Alloc >::data(), Vector< T, Alloc >::device_data(), BLAS::gemv(), qmcplusplus::ompBLAS::gemv_batched(), BLAS::gemv_trans(), qmcplusplus::Units::force::N, Vector< T, Alloc >::resize(), and Vector< T, Alloc >::updateTo().
| void qmcplusplus::test_gemv_cases | ( | ) |
| void qmcplusplus::test_ger | ( | const int | M, |
| const int | N | ||
| ) |
Definition at line 393 of file test_ompBLAS.cpp.
References CHECK(), TinyVector< T, D >::data(), qmcplusplus::ompBLAS::ger(), BLAS::ger(), and qmcplusplus::Units::force::N.
| void test_ger_batched | ( | const int | M, |
| const int | N, | ||
| const int | batch_count | ||
| ) |
Definition at line 436 of file test_ompBLAS.cpp.
References CHECK(), Vector< T, Alloc >::data(), Vector< T, Alloc >::device_data(), BLAS::ger(), qmcplusplus::ompBLAS::ger_batched(), qmcplusplus::Units::force::N, Vector< T, Alloc >::resize(), and Vector< T, Alloc >::updateTo().
| void qmcplusplus::test_ger_cases | ( | ) |
| void qmcplusplus::test_HCN | ( | bool | transform | ) |
Definition at line 595 of file test_MO.cpp.
References castCharToXMLChar(), CHECK(), OHMMS::Controller, SPOSetBuilderFactory::createSPOSetBuilder(), doc, qmcplusplus::Units::charge::e, Libxml2Document::getRoot(), Libxml2Document::getXPathContext(), okay, Libxml2Document::parse(), XMLParticleParser::readXML(), and REQUIRE().
Referenced by TEST_CASE().
| void qmcplusplus::test_hcpBe_rotation | ( | bool | use_single_det, |
| bool | use_nlpp_batched | ||
| ) |
Definition at line 38 of file test_RotatedSPOs_NLPP.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), ProjectData::BATCH, CHECK(), TrialWaveFunction::checkInVariables(), TrialWaveFunction::checkOutVariables(), OHMMS::Controller, doc, qmcplusplus::Units::charge::e, OperatorBase::evaluate(), QMCHamiltonian::evaluate(), TrialWaveFunction::evaluateDerivatives(), TrialWaveFunction::evaluateLog(), QMCHamiltonian::evaluateValueAndDerivatives(), QMCHamiltonian::getHamiltonian(), QMCHamiltonian::getKineticEnergy(), QMCHamiltonian::getLocalEnergy(), TrialWaveFunction::getOrbitals(), ParticleSetPool::getPool(), Libxml2Document::getRoot(), ProjectData::getRuntimeOptions(), ParticleSetPool::getSimulationCell(), lattice, TrialWaveFunction::mw_evaluateParameterDerivatives(), QMCHamiltonian::mw_evaluateValueAndDerivatives(), okay, Libxml2Document::parseFromString(), REQUIRE(), VariableSet::resetIndex(), TrialWaveFunction::resetParameters(), QMCHamiltonian::setRandomGenerator(), ParticleSetPool::setSimulationCell(), and test_project.
Referenced by TEST_CASE().
| void qmcplusplus::test_He | ( | bool | transform | ) |
Definition at line 26 of file test_MO.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), castCharToXMLChar(), CHECK(), OHMMS::Controller, SPOSetBuilderFactory::createSPOSetBuilder(), doc, Libxml2Document::getRoot(), Libxml2Document::getXPathContext(), ispecies, okay, Libxml2Document::parse(), and REQUIRE().
Referenced by TEST_CASE().
| void qmcplusplus::test_He_mw | ( | bool | transform | ) |
Definition at line 136 of file test_MO.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), castCharToXMLChar(), CHECK(), OHMMS::Controller, SPOSetBuilderFactory::createSPOSetBuilder(), doc, Libxml2Document::getRoot(), Libxml2Document::getXPathContext(), ispecies, ParticleSet::makeMove(), okay, Libxml2Document::parse(), ParticleSet::R, REQUIRE(), and ParticleSet::update().
Referenced by TEST_CASE().
| void qmcplusplus::test_He_sto3g_xml_input | ( | const std::string & | spo_xml_string | ) |
Definition at line 30 of file test_spo_collection_input_LCAO_xml.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), CHECK(), OHMMS::Controller, ParticleSet::create(), doc, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), ispecies, okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), ParticleSet::setName(), and ParticleSet::update().
Referenced by TEST_CASE().
| void qmcplusplus::test_inverse | ( | const std::int64_t | N | ) |
Definition at line 24 of file test_syclSolverInverter.cpp.
References CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), getSYCLDefaultDeviceDefaultQueue(), syclSolverInverter< T_FP >::invert_transpose(), DiracMatrix< T_FP >::invert_transpose(), qmcplusplus::Units::distance::m, qmcplusplus::testing::makeRngSpdMatrix(), qmcplusplus::Units::force::N, queue, and Matrix< T, Alloc >::resize().
| void qmcplusplus::test_isnan | ( | ) |
Definition at line 22 of file test_math.cpp.
References CHECK(), isnan(), and sqrt().
| void qmcplusplus::test_J1_spline | ( | const DynamicCoordinateKind | kind_selected | ) |
Definition at line 61 of file test_J1_bspline.cpp.
References ParticleSet::acceptMove(), SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), RadialJastrowBuilder::buildComponent(), CHECK(), OHMMS::Controller, ParticleSet::create(), ParticleSet::createResource(), BsplineFunctor< REAL >::CuspValue, DC_POS_OFFLOAD, BsplineFunctor< REAL >::DeltaR, doc, BsplineFunctor< REAL >::evaluate(), ParticleSet::G, ParticleSet::getDistTableAB(), Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), ParticleSet::getTotalNum(), ispecies, ParticleSet::L, qmcplusplus::Units::distance::m, ParticleSet::makeMove(), ParticleSet::makeVirtualMoves(), VirtualParticleSet::mw_makeMoves(), ParticleSet::mw_update(), qmcplusplus::Units::force::N, n, BsplineFunctor< REAL >::NumParams, OHMMS_DIM, okay, Libxml2Document::parseFromString(), ParticleSet::R, ParticleSet::rejectMove(), REQUIRE(), ParticleSet::resetGroups(), ParticleSet::setName(), and ParticleSet::update().
Referenced by TEST_CASE().
| void qmcplusplus::test_J3_polynomial3D | ( | const DynamicCoordinateKind | kind_selected | ) |
Definition at line 46 of file test_polynomial_eeI_jastrow.cpp.
References SpeciesSet::addSpecies(), ParticleSet::addTable(), app_log(), eeI_JastrowBuilder::buildComponent(), CHECK(), WaveFunctionComponent::checkOutVariables(), OHMMS::Controller, ParticleSet::create(), WaveFunctionComponent::createResource(), ParticleSet::createResource(), doc, Dot(), qmcplusplus::Units::charge::e, WaveFunctionComponent::evaluateDerivatives(), WaveFunctionComponent::evaluateDerivativesWF(), WaveFunctionComponent::evaluateDerivRatios(), WaveFunctionComponent::evaluateLog(), WaveFunctionComponent::evaluateRatios(), WaveFunctionComponent::evaluateRatiosAlltoOne(), WaveFunctionComponent::extractOptimizableObjectRefs(), ParticleSet::G, ParticleSet::getDistTableAB(), Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), ParticleSet::getTotalNum(), ParticleSet::L, WaveFunctionComponent::makeClone(), ParticleSet::makeMove(), VirtualParticleSet::makeMoves(), ParticleSet::makeVirtualMoves(), WaveFunctionComponent::mw_evaluateRatios(), VirtualParticleSet::mw_makeMoves(), WaveFunctionComponent::mw_recompute(), ParticleSet::mw_update(), okay, Libxml2Document::parseFromString(), ParticleSet::R, WaveFunctionComponent::ratio(), ParticleSet::rejectMove(), REQUIRE(), Vector< T, Alloc >::resize(), ParticleSet::setName(), Sum(), and ParticleSet::update().
Referenced by TEST_CASE().
| void qmcplusplus::test_LCAO_DiamondC_2x1x1_cplx | ( | const bool | useOffload | ) |
Definition at line 454 of file test_LCAO_diamondC_2x1x1.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), app_log(), CHECK(), OHMMS::Controller, ParticleSet::create(), VirtualParticleSet::createResource(), ParticleSet::createResource(), LCAOrbitalBuilder::createSPOSetFromXML(), QMCTraits::DIM, doc, ParticleSet::getDistTableAB(), Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), imag(), ispecies, lattice, VirtualParticleSet::mw_makeMoves(), ParticleSet::mw_update(), okay, Libxml2Document::parseFromString(), ParticleSet::print(), LatticeParser::put(), ParticleSet::R, REQUIRE(), ParticleSet::setName(), and ParticleSet::update().
Referenced by TEST_CASE().
| void qmcplusplus::test_LCAO_DiamondC_2x1x1_real | ( | const bool | useOffload | ) |
Definition at line 37 of file test_LCAO_diamondC_2x1x1.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), app_log(), CHECK(), OHMMS::Controller, ParticleSet::create(), VirtualParticleSet::createResource(), ParticleSet::createResource(), LCAOrbitalBuilder::createSPOSetFromXML(), Vector< T, Alloc >::data(), Vector< T, Alloc >::device_data(), QMCTraits::DIM, QMCTraits::DIM_VGL, doc, ParticleSet::getDistTableAB(), Libxml2Document::getRoot(), ParticleSet::getSpeciesSet(), imag(), ispecies, lattice, VirtualParticleSet::mw_makeMoves(), ParticleSet::mw_update(), okay, Libxml2Document::parseFromString(), ParticleSet::print(), LatticeParser::put(), ParticleSet::R, REQUIRE(), Array< T, D, ALLOC >::resize(), ParticleSet::setName(), ParticleSet::update(), and Vector< T, Alloc >::updateTo().
Referenced by TEST_CASE().
| void qmcplusplus::test_lcao_spinor | ( | ) |
this is a somewhat simple example. We have an ion at the origin and a gaussian basis function centered on the ion as a orbital. In this case, the ion derivative is actually just the negative of the electron gradient.
Definition at line 26 of file test_MO_spinor.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), ParticleSet::addTable(), app_log(), CHECK(), OHMMS::Controller, ParticleSet::create(), ParticleSet::createResource(), SPOSetBuilderFactory::createSPOSetBuilder(), doc, qmcplusplus::Units::charge::e, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), ispecies, SpinorSet::isResourceOwned(), ParticleSet::makeMove(), ParticleSet::mw_accept_rejectMove(), ParticleSet::mw_makeMove(), ParticleSet::mw_update(), okay, Libxml2Document::parseFromString(), POS_SPIN, MCCoords< CoordsType::POS_SPIN >::positions, processChildren(), ParticleSet::R, ParticleSet::rejectMove(), REQUIRE(), Matrix< T, Alloc >::resize(), Vector< T, Alloc >::resize(), ParticleSet::setName(), ParticleSet::setSpinor(), MCCoords< CoordsType::POS_SPIN >::spins, ParticleSet::spins, and ParticleSet::update().
Referenced by TEST_CASE().
| void qmcplusplus::test_lcao_spinor_excited | ( | ) |
this is a somewhat simple example. We have an ion at the origin and a gaussian basis function centered on the ion as a orbital. In this case, the ion derivative is actually just the negative of the electron gradient.
Definition at line 353 of file test_MO_spinor.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), ParticleSet::addTable(), app_log(), CHECK(), OHMMS::Controller, ParticleSet::create(), ParticleSet::createResource(), SPOSetBuilderFactory::createSPOSetBuilder(), doc, qmcplusplus::Units::charge::e, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), ispecies, ParticleSet::makeMove(), ParticleSet::mw_accept_rejectMove(), ParticleSet::mw_makeMove(), ParticleSet::mw_update(), okay, Libxml2Document::parseFromString(), POS_SPIN, MCCoords< CoordsType::POS_SPIN >::positions, processChildren(), ParticleSet::R, ParticleSet::rejectMove(), REQUIRE(), Matrix< T, Alloc >::resize(), Vector< T, Alloc >::resize(), ParticleSet::setName(), ParticleSet::setSpinor(), MCCoords< CoordsType::POS_SPIN >::spins, ParticleSet::spins, and ParticleSet::update().
Referenced by TEST_CASE().
| void qmcplusplus::test_lcao_spinor_ion_derivs | ( | ) |
Definition at line 676 of file test_MO_spinor.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), ParticleSet::addTable(), app_log(), CHECK(), OHMMS::Controller, ParticleSet::create(), SPOSetBuilderFactory::createSPOSetBuilder(), doc, qmcplusplus::Units::charge::e, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), ispecies, okay, Libxml2Document::parseFromString(), processChildren(), ParticleSet::R, REQUIRE(), ParticleSet::setName(), ParticleSet::setSpinor(), ParticleSet::spins, and ParticleSet::update().
Referenced by TEST_CASE().
| void qmcplusplus::test_LiH_msd | ( | const std::string & | spo_xml_string, |
| const std::string & | check_sponame, | ||
| int | check_spo_size, | ||
| int | check_basisset_size, | ||
| int | test_nlpp_algorithm_batched, | ||
| int | test_batched_api | ||
| ) |
Definition at line 40 of file test_multi_slater_determinant.cpp.
References ParticleSet::acceptMove(), SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), app_log(), CHECK(), OHMMS::Controller, ParticleSet::create(), ParticleSet::createResource(), doc, ParticleSet::G, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), TWFGrads< CoordsType::POS >::grads_positions, ispecies, ParticleSet::L, ParticleSet::makeMove(), VirtualParticleSet::makeMoves(), ParticleSet::mw_accept_rejectMove(), TrialWaveFunction::mw_accept_rejectMove(), TrialWaveFunction::mw_calcRatio(), TrialWaveFunction::mw_calcRatioGrad(), TrialWaveFunction::mw_evalGrad(), TrialWaveFunction::mw_evaluateLog(), ParticleSet::mw_makeMove(), TrialWaveFunction::mw_prepareGroup(), ParticleSet::mw_update(), okay, Libxml2Document::parseFromString(), ParticleSet::R, ParticleSet::rejectMove(), REQUIRE(), ParticleSet::resetGroups(), ParticleSet::setName(), VariableSet::size_of_active(), twf, and ParticleSet::update().
Referenced by TEST_CASE().
| void qmcplusplus::test_LiH_msd_xml_input | ( | const std::string & | spo_xml_string, |
| const std::string & | check_sponame, | ||
| int | check_spo_size, | ||
| int | check_basisset_size | ||
| ) |
Definition at line 31 of file test_spo_collection_input_MSD_LCAO_h5.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), OHMMS::Controller, ParticleSet::create(), doc, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), ispecies, okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), and ParticleSet::setName().
Referenced by TEST_CASE().
| void qmcplusplus::test_LiH_msd_xml_input_with_positron | ( | const std::string & | spo_xml_string, |
| const std::string & | check_sponame, | ||
| int | check_spo_size, | ||
| int | check_basisset_size | ||
| ) |
Definition at line 194 of file test_spo_collection_input_MSD_LCAO_h5.cpp.
References SpeciesSet::addAttribute(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), OHMMS::Controller, ParticleSet::create(), doc, ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), ispecies, okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), and ParticleSet::setName().
Referenced by TEST_CASE().
| void qmcplusplus::test_Ne | ( | bool | transform | ) |
Definition at line 465 of file test_MO.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), castCharToXMLChar(), CHECK(), OHMMS::Controller, SPOSetBuilderFactory::createSPOSetBuilder(), doc, Libxml2Document::getRoot(), Libxml2Document::getXPathContext(), ispecies, VirtualParticleSet::makeMoves(), okay, Libxml2Document::parse(), and REQUIRE().
Referenced by TEST_CASE().
| void qmcplusplus::test_one_gemm | ( | const int | M, |
| const int | N, | ||
| const int | K, | ||
| const char | transa, | ||
| const char | transb | ||
| ) |
Definition at line 23 of file test_AccelBLAS.cpp.
References qmcplusplus::Units::distance::A, B(), qmcplusplus::Units::charge::C, CHECK(), Vector< T, Alloc >::device_data(), qmcplusplus::compute::BLAS::gemm(), BLAS::gemm(), qmcplusplus::compute::BLAS::gemm_batched(), imag(), qmcplusplus::Units::energy::K, qmcplusplus::Units::force::N, queue, and Vector< T, Alloc >::updateTo().
| void qmcplusplus::test_one_gemv | ( | const int | M_b, |
| const int | N_b, | ||
| const char | trans | ||
| ) |
Definition at line 177 of file test_AccelBLAS.cpp.
References qmcplusplus::Units::distance::A, B(), qmcplusplus::Units::charge::C, CHECK(), TinyVector< T, D >::data(), Vector< T, Alloc >::device_data(), BLAS::gemv(), qmcplusplus::compute::BLAS::gemv(), qmcplusplus::compute::BLAS::gemv_batched(), BLAS::gemv_trans(), qmcplusplus::Units::force::N, queue, and Vector< T, Alloc >::updateTo().
| void qmcplusplus::test_one_ger | ( | const int | M, |
| const int | N | ||
| ) |
Definition at line 317 of file test_AccelBLAS.cpp.
References CHECK(), TinyVector< T, D >::data(), Vector< T, Alloc >::device_data(), qmcplusplus::compute::BLAS::ger(), BLAS::ger(), qmcplusplus::compute::BLAS::ger_batched(), qmcplusplus::Units::force::N, queue, and Vector< T, Alloc >::updateTo().
| void qmcplusplus::test_real_accumulator | ( | ) |
Definition at line 59 of file test_accumulator.cpp.
References CHECK(), accumulator_set< T, typename >::count(), accumulator_set< T, typename >::mean(), mean(), accumulator_set< T, typename >::mean2(), accumulator_set< T, typename >::mean_and_variance(), accumulator_set< T, typename >::result(), accumulator_set< T, typename >::result2(), and accumulator_set< T, typename >::variance().
| void qmcplusplus::test_real_accumulator_basic | ( | ) |
Definition at line 26 of file test_accumulator.cpp.
References accumulator_set< T, typename >::bad(), CHECK(), accumulator_set< T, typename >::count(), accumulator_set< T, typename >::good(), accumulator_set< T, typename >::mean(), accumulator_set< T, typename >::mean2(), REQUIRE(), and accumulator_set< T, typename >::variance().
| void qmcplusplus::test_real_accumulator_weights | ( | ) |
Definition at line 90 of file test_accumulator.cpp.
References CHECK(), accumulator_set< T, typename >::count(), accumulator_set< T, typename >::mean(), accumulator_set< T, typename >::mean2(), accumulator_set< T, typename >::mean_and_variance(), accumulator_set< T, typename >::result(), accumulator_set< T, typename >::result2(), and accumulator_set< T, typename >::variance().
| void qmcplusplus::test_spline_bounds | ( | ) |
Definition at line 19 of file test_SplineBound.cpp.
References CHECK(), getSplineBound(), and REQUIRE().
| void qmcplusplus::test_tiny_vector | ( | ) |
Definition at line 28 of file test_TinyVector.cpp.
References CHECK(), dot(), and REQUIRE().
| void qmcplusplus::test_tiny_vector_size_two | ( | ) |
Definition at line 65 of file test_TinyVector.cpp.
References CHECK().
| void qmcplusplus::testDualAllocator | ( | ) |
Definition at line 40 of file test_dual_allocators_ohmms_containers.cpp.
References CHECK(), check_matrix_result, CHECKED_ELSE(), checkMatrix(), qmcplusplus::syclBLAS::copy_n(), VectorSoaContainer< T, D, Alloc >::copyDeviceDataByIndex(), Matrix< T, Alloc >::data(), VectorSoaContainer< T, D, Alloc >::data(), Matrix< T, Alloc >::device_data(), VectorSoaContainer< T, D, Alloc >::device_data(), qmcplusplus::testing::makeRngSpdMatrix(), n, Array< T, D, ALLOC >::resize(), Matrix< T, Alloc >::resize(), VectorSoaContainer< T, D, Alloc >::resize(), Array< T, D, ALLOC >::size(), VectorSoaContainer< T, D, Alloc >::updateFrom(), and VectorSoaContainer< T, D, Alloc >::updateTo().
| void qmcplusplus::testElecCase | ( | double | mass_up, |
| double | mass_dn | ||
| ) |
Provide a test scope parameterized on electron species mass that then can run a set of tests using its objects.
I couldn't see a easier solution to deal with all the setup to get both coverage of this interesting feature considering various approaches to copy constructors the objects in the testing scope take.
Definition at line 230 of file test_bare_kinetic.cpp.
References SpeciesSet::addAttribute(), SpeciesSet::addSpecies(), ParticleSet::addTable(), ParticleSet::create(), BareKineticEnergy::createResource(), ParticleSet::createResource(), qmcplusplus::testing::getParticularListener(), ParticleSet::getSpeciesSet(), ParticleSet::L, ParticleSet::R, ParticleSet::setName(), and ParticleSet::update().
Referenced by TEST_CASE().
| embt qmcplusplus::testMakeBlockAverages | ( | ) |
| embt qmcplusplus::testReduceOperatorEstimators | ( | ) |
| void qmcplusplus::TestTask | ( | const int | ip, |
| std::atomic< int > & | counter | ||
| ) |
Definition at line 22 of file test_ParallelExecutorSTD.cpp.
Referenced by TEST_CASE().
| void qmcplusplus::TestTaskOMP | ( | const int | ip, |
| int & | counter | ||
| ) |
Openmp generally works but is not guaranteed with std::atomic.
Definition at line 20 of file test_ParallelExecutorOPENMP.cpp.
Referenced by TEST_CASE().
| void qmcplusplus::testTrialWaveFunction_diamondC_2x1x1 | ( | const int | ndelay, |
| const OffloadSwitches & | offload_switches | ||
| ) |
Templated test of TrialWF with different DiracDet flavors.
Definition at line 51 of file test_TrialWaveFunction_diamondC_2x1x1.cpp.
References TrialWaveFunction::acceptMove(), SpeciesSet::addAttribute(), TrialWaveFunction::addComponent(), ParticleSetPool::addParticleSet(), SpeciesSet::addSpecies(), ParticleSet::addTable(), app_log(), RadialJastrowBuilder::buildComponent(), TrialWaveFunction::calcRatio(), CHECK(), OHMMS::Controller, ParticleSet::create(), TrialWaveFunction::createResource(), ParticleSet::createSK(), EinsplineSetBuilder::createSPOSetFromXML(), DC_POS, DC_POS_OFFLOAD, doc, qmcplusplus::Units::charge::e, TrialWaveFunction::evaluateGL(), TrialWaveFunction::evaluateLog(), TrialWaveFunction::FERMIONIC, ParticleSet::getDistTableAB(), TrialWaveFunction::getLogPsi(), TrialWaveFunction::getPhase(), ParticleSetPool::getPool(), Libxml2Document::getRoot(), ParticleSetPool::getSimulationCell(), ParticleSet::getSpeciesSet(), TWFGrads< CoordsType::POS >::grads_positions, OffloadSwitches::jas, lattice, log_values(), TrialWaveFunction::makeClone(), ParticleSet::mw_accept_rejectMove(), TrialWaveFunction::mw_accept_rejectMove(), TrialWaveFunction::mw_calcRatio(), TrialWaveFunction::mw_calcRatioGrad(), TrialWaveFunction::mw_completeUpdates(), TrialWaveFunction::mw_evalGrad(), TrialWaveFunction::mw_evaluateGL(), TrialWaveFunction::mw_evaluateLog(), TrialWaveFunction::mw_evaluateRatios(), ParticleSet::mw_makeMove(), VirtualParticleSet::mw_makeMoves(), ParticleSet::mw_update(), TrialWaveFunction::NONFERMIONIC, okay, Libxml2Document::parseFromString(), ParticleSet::R, REQUIRE(), ParticleSet::resetGroups(), ParticleSet::setName(), ParticleSetPool::setSimulationCell(), OffloadSwitches::spo, and ParticleSet::update().
| T qmcplusplus::trace | ( | const SymTensor< T, D > & | rhs | ) |
Definition at line 321 of file SymTensor.h.
|
inline |
| T qmcplusplus::trace | ( | const AntiSymTensor< T, D > & | ) |
Definition at line 468 of file AntiSymTensor.h.
Referenced by CountingGaussian::evaluate(), CountingGaussian::evaluate_print(), and CountingGaussian::evaluateLog().
Tr(a^t *b), 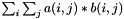 .
.
Definition at line 361 of file Tensor.h.
Referenced by DiracDeterminantWithBackflow::dummyEvalLi(), DiracDeterminantWithBackflow::evaluateDerivatives(), and DiracDeterminantWithBackflow::evaluateLog().
|
inline |
| bool qmcplusplus::TraceSample_comp | ( | TraceSample< T > * | left, |
| TraceSample< T > * | right | ||
| ) |
Definition at line 744 of file TraceManager.h.
References TraceSample< T >::data_size.
Definition at line 330 of file SymTensor.h.
| AntiSymTensor<T, D> qmcplusplus::transpose | ( | const AntiSymTensor< T, D > & | rhs | ) |
Definition at line 474 of file AntiSymTensor.h.
Referenced by BsplineReader::check_twists(), SplineR2R< ST >::create_spline(), DiracDeterminantWithBackflow::dummyEvalLi(), DiracDeterminantWithBackflow::evaluateDerivatives(), DiracDeterminantWithBackflow::evaluateLog(), qmcplusplus::ewaldref::ewaldSum(), qmcplusplus::ewaldref::madelungSum(), CrystalLattice< ST, 3 >::reset(), and EinsplineSetBuilder::set_metadata().
| template void qmcplusplus::TrialWaveFunction::mw_calcRatioGrad< CoordsType::POS > | ( | const RefVectorWithLeader< TrialWaveFunction > & | wf_list, |
| const RefVectorWithLeader< ParticleSet > & | p_list, | ||
| int | iat, | ||
| std::vector< PsiValue > & | ratios, | ||
| TWFGrads< CoordsType::POS > & | grads | ||
| ) |
| template void qmcplusplus::TrialWaveFunction::mw_calcRatioGrad< CoordsType::POS_SPIN > | ( | const RefVectorWithLeader< TrialWaveFunction > & | wf_list, |
| const RefVectorWithLeader< ParticleSet > & | p_list, | ||
| int | iat, | ||
| std::vector< PsiValue > & | ratios, | ||
| TWFGrads< CoordsType::POS_SPIN > & | grads | ||
| ) |
| template void qmcplusplus::TrialWaveFunction::mw_evalGrad< CoordsType::POS > | ( | const RefVectorWithLeader< TrialWaveFunction > & | wf_list, |
| const RefVectorWithLeader< ParticleSet > & | p_list, | ||
| int | iat, | ||
| TWFGrads< CoordsType::POS > & | grads | ||
| ) |
| template void qmcplusplus::TrialWaveFunction::mw_evalGrad< CoordsType::POS_SPIN > | ( | const RefVectorWithLeader< TrialWaveFunction > & | wf_list, |
| const RefVectorWithLeader< ParticleSet > & | p_list, | ||
| int | iat, | ||
| TWFGrads< CoordsType::POS_SPIN > & | grads | ||
| ) |
| template void qmcplusplus::TWFdispatcher::flex_calcRatioGrad< CoordsType::POS > | ( | const RefVectorWithLeader< TrialWaveFunction > & | wf_list, |
| const RefVectorWithLeader< ParticleSet > & | p_list, | ||
| int | iat, | ||
| std::vector< PsiValue > & | ratios, | ||
| TWFGrads< CoordsType::POS > & | grads | ||
| ) | const |
| template void qmcplusplus::TWFdispatcher::flex_calcRatioGrad< CoordsType::POS_SPIN > | ( | const RefVectorWithLeader< TrialWaveFunction > & | wf_list, |
| const RefVectorWithLeader< ParticleSet > & | p_list, | ||
| int | iat, | ||
| std::vector< PsiValue > & | ratios, | ||
| TWFGrads< CoordsType::POS_SPIN > & | grads | ||
| ) | const |
| template void qmcplusplus::TWFdispatcher::flex_evalGrad< CoordsType::POS > | ( | const RefVectorWithLeader< TrialWaveFunction > & | wf_list, |
| const RefVectorWithLeader< ParticleSet > & | p_list, | ||
| int | iat, | ||
| TWFGrads< CoordsType::POS > & | grads | ||
| ) | const |
| template void qmcplusplus::TWFdispatcher::flex_evalGrad< CoordsType::POS_SPIN > | ( | const RefVectorWithLeader< TrialWaveFunction > & | wf_list, |
| const RefVectorWithLeader< ParticleSet > & | p_list, | ||
| int | iat, | ||
| TWFGrads< CoordsType::POS_SPIN > & | grads | ||
| ) | const |
|
inline |
unpack packed cG to fftbox
| cG | packed vector |
| gvecs | g-coordinate for cG[i] |
| maxg | fft grid |
| fftbox | unpacked data to be transformed |
Definition at line 34 of file einspline_helper.hpp.
References lower_bound(), and upper_bound().
Referenced by OneSplineOrbData::fft_spline().
|
inline |
helper function to determine the upper bound of a domain (need to move up)
Definition at line 208 of file InitMolecularSystem.cpp.
Referenced by HEGGrid< T >::getShellIndex(), InitMolecularSystem::initWithVolume(), and unpack4fftw().
| T qmcplusplus::v_m_v | ( | T | h00, |
| T | h01, | ||
| T | h02, | ||
| T | h11, | ||
| T | h12, | ||
| T | h22, | ||
| T | g1x, | ||
| T | g1y, | ||
| T | g1z, | ||
| T | g2x, | ||
| T | g2y, | ||
| T | g2z | ||
| ) |
compute vector[3]^T x matrix[3][3] x vector[3]
Definition at line 54 of file contraction_helper.hpp.
Referenced by SplineC2R< ST >::assign_vgh(), SplineR2R< ST >::assign_vgh(), SplineC2C< ST >::assign_vgh(), SplineC2COMPTarget< ST >::assign_vgh(), SplineC2ROMPTarget< ST >::assign_vgh(), SplineC2R< ST >::assign_vghgh(), SplineR2R< ST >::assign_vghgh(), SplineC2C< ST >::assign_vghgh(), SplineC2COMPTarget< ST >::assign_vghgh(), and SplineC2ROMPTarget< ST >::assign_vghgh().
| template void qmcplusplus::VMCBatched::advanceWalkers< CoordsType::POS > | ( | const StateForThread & | sft, |
| Crowd & | crowd, | ||
| QMCDriverNew::DriverTimers & | timers, | ||
| ContextForSteps & | step_context, | ||
| bool | recompute, | ||
| bool | accumulate_this_step | ||
| ) |
| template void qmcplusplus::VMCBatched::advanceWalkers< CoordsType::POS_SPIN > | ( | const StateForThread & | sft, |
| Crowd & | crowd, | ||
| QMCDriverNew::DriverTimers & | timers, | ||
| ContextForSteps & | step_context, | ||
| bool | recompute, | ||
| bool | accumulate_this_step | ||
| ) |
| template void qmcplusplus::WaveFunctionComponent::mw_evalGrad< CoordsType::POS > | ( | const RefVectorWithLeader< WaveFunctionComponent > & | wfc_list, |
| const RefVectorWithLeader< ParticleSet > & | p_list, | ||
| int | iat, | ||
| TWFGrads< CoordsType::POS > & | grad_now | ||
| ) | const |
| template void qmcplusplus::WaveFunctionComponent::mw_evalGrad< CoordsType::POS_SPIN > | ( | const RefVectorWithLeader< WaveFunctionComponent > & | wfc_list, |
| const RefVectorWithLeader< ParticleSet > & | p_list, | ||
| int | iat, | ||
| TWFGrads< CoordsType::POS_SPIN > & | grad_now | ||
| ) | const |
| template void qmcplusplus::WaveFunctionComponent::mw_ratioGrad< CoordsType::POS > | ( | const RefVectorWithLeader< WaveFunctionComponent > & | wfc_list, |
| const RefVectorWithLeader< ParticleSet > & | p_list, | ||
| int | iat, | ||
| std::vector< PsiValue > & | ratios, | ||
| TWFGrads< CoordsType::POS > & | grad_new | ||
| ) | const |
| template void qmcplusplus::WaveFunctionComponent::mw_ratioGrad< CoordsType::POS_SPIN > | ( | const RefVectorWithLeader< WaveFunctionComponent > & | wfc_list, |
| const RefVectorWithLeader< ParticleSet > & | p_list, | ||
| int | iat, | ||
| std::vector< PsiValue > & | ratios, | ||
| TWFGrads< CoordsType::POS_SPIN > & | grad_new | ||
| ) | const |
|
inline |
Definition at line 454 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 487 of file ParticleAttrib.cpp.
References MakeReturn< T >::make().
|
inline |
Definition at line 725 of file OhmmsMatrixOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 1367 of file OhmmsVectorOperators.h.
References MakeReturn< T >::make().
|
inline |
Definition at line 54 of file string_utils.h.
Referenced by split().
| void qmcplusplus::wrapAroundBox | ( | const PL & | lat, |
| const PV & | pin, | ||
| PV & | pout | ||
| ) |
Definition at line 78 of file ParticleUtility.h.
| void X2alpha | ( | const TinyVector< ValueType, 5 > & | X, |
| RealType | Rc, | ||
| TinyVector< ValueType, 5 > & | alpha | ||
| ) |
Convert constraints to polynomial parameters.
| X | input from evalX |
| Rc | cutoff radius |
| alpha | output the polynomial parameters for the correction |
Definition at line 348 of file CuspCorrectionConstruction.cpp.
Referenced by evaluateForPhi0Body().
|
inline |
wrappers around xgetrf lapack routines
| [in] | n | rows |
| [in] | m | cols |
| [in,out] | a | matrix contains LU matrix after call |
| [in] | lda | leading dimension of a |
| [out] | piv | pivot vector |
Definition at line 31 of file DiracMatrix.h.
References lda, qmcplusplus::Units::distance::m, n, and sgetrf().
Referenced by DiracMatrixComputeOMPTarget< VALUE_FP >::computeInvertAndLog(), DiracMatrix< VALUE_FP >::computeInvertAndLog(), and TEST_CASE().
|
inline |
Definition at line 38 of file DiracMatrix.h.
References cgetrf(), lda, qmcplusplus::Units::distance::m, and n.
|
inline |
Definition at line 45 of file DiracMatrix.h.
References dgetrf(), lda, qmcplusplus::Units::distance::m, and n.
|
inline |
Definition at line 52 of file DiracMatrix.h.
References lda, qmcplusplus::Units::distance::m, n, and zgetrf().
|
inline |
inversion of a float matrix after lu factorization
Definition at line 60 of file DiracMatrix.h.
References lda, n, and sgetri().
Referenced by DiracMatrixComputeOMPTarget< VALUE_FP >::computeInvertAndLog(), DiracMatrix< VALUE_FP >::computeInvertAndLog(), DiracMatrixComputeOMPTarget< VALUE_FP >::reset(), and DiracMatrix< VALUE_FP >::reset().
|
inline |
Definition at line 67 of file DiracMatrix.h.
References cgetri(), lda, and n.
|
inline |
Definition at line 79 of file DiracMatrix.h.
References dgetri(), lda, and n.
|
inline |
inversion of a std::complex<double> matrix after lu factorization
Definition at line 87 of file DiracMatrix.h.
References lda, n, and zgetri().
|
inline |
calculates Ylm param[in] l angular momentum param[in] m magnetic quantum number param[in] r position vector.
Note: This must be a unit vector and in the order [z,x,y] to be correct
Definition at line 89 of file Ylm.h.
References atan2(), cos(), LegendrePlm(), qmcplusplus::Units::distance::m, sin(), and sqrt().
Referenced by SoaSphericalTensor< ST >::batched_evaluateV(), Gvectors< ST, LT >::calc_Ylm_G(), Quadrature3D< T >::CheckQuadratureRule(), Quadrature3D< T >::CheckQuadratureRuleReal(), HybridRepSetReader< SA >::create_atomic_centers_Gspace(), derivYlmSpherical(), SoaSphericalTensor< ST >::evaluateV(), sphericalHarmonic(), and TEST_CASE().
|
static |
Definition at line 21 of file test_output_manager.cpp.
Referenced by init_string_output(), reset_string_output(), and TEST_CASE().
| int batch_size = 2 |
Definition at line 219 of file test_cuBLAS_LU.cpp.
Referenced by compute_batch_parameters(), DiracMatrixComputeOMPTarget< VALUE_FP >::reset(), and TEST_CASE().
| check_matrix_result = checkMatrix(lu_mat, M_mat) |
Definition at line 454 of file test_cuBLAS_LU.cpp.
Referenced by CHECKED_ELSE(), TEMPLATE_TEST_CASE(), TEST_CASE(), test_inverse(), and testDualAllocator().
| auto checkArray |
Definition at line 441 of file test_cuBLAS_LU.cpp.
Referenced by TestFillBufferRngReal< T >::operator()(), TestFillVecRngReal< T >::operator()(), TestGetVecRngReal< T >::operator()(), TestFillBufferRngComplex< T >::operator()(), TestFillVecRngComplex< T >::operator()(), TestGetVecRngComplex< T >::operator()(), and TEST_CASE().
| auto checkRs |
Definition at line 117 of file test_random_seq.cpp.
| auto clone = original.spawnCrowdClone() |
Definition at line 376 of file test_OneBodyDensityMatrices.cpp.
Referenced by Backflow_eI< FT >::makeClone(), Backflow_ee< FT >::makeClone(), Backflow_ee_kSpace::makeClone(), Backflow_eI_spin< FT >::makeClone(), BackflowTransformation::makeClone(), MultiSlaterDetTableMethod::makeClone(), OrbitalImages::makeClone(), TEST_CASE(), and h5data_proxy< std::ostringstream >::write().
| const std::string coeff_rot_by_point05 |
Definition at line 534 of file test_RotatedSPOs_LCAO.cpp.
Referenced by TEST_CASE().
| const std::string coeff_rot_by_point1 |
Definition at line 465 of file test_RotatedSPOs_LCAO.cpp.
Referenced by TEST_CASE().
| const std::string coeff_rot_by_point2 |
Definition at line 470 of file test_RotatedSPOs_LCAO.cpp.
Referenced by TEST_CASE().
| Communicate * comm = OHMMS::Controller |
Definition at line 50 of file test_EstimatorManagerNew.cpp.
Referenced by QMCDriverNew::adjustGlobalWalkerCount(), HybridRepCplx< SPLINEBASE >::bcast_tables(), HybridRepReal< SPLINEBASE >::bcast_tables(), AtomicOrbitals< ST >::bcast_tables(), SplineC2R< ST >::bcast_tables(), SplineR2R< ST >::bcast_tables(), SplineC2C< ST >::bcast_tables(), SplineC2COMPTarget< ST >::bcast_tables(), SplineC2ROMPTarget< ST >::bcast_tables(), HybridRepCenterOrbitals< SPLINEBASE::DataType >::bcast_tables(), TimerManager< qmcplusplus::TimerType< CLOCK > >::collate_flat_profile(), DMCFactory::create(), RMCFactory::create(), VMCFactory::create(), DMCFactoryNew::create(), VMCFactoryNew::create(), qmcplusplus::testing::createDriver(), QMCDriverFactory::createQMCDriver(), createWalkerController(), fill_from_text(), HybridRepCenterOrbitals< SPLINEBASE::DataType >::gather_atomic_tables(), HybridRepCplx< SPLINEBASE >::gather_tables(), HybridRepReal< SPLINEBASE >::gather_tables(), AtomicOrbitals< ST >::gather_tables(), SplineC2R< ST >::gather_tables(), SplineR2R< ST >::gather_tables(), SplineC2C< ST >::gather_tables(), SplineC2COMPTarget< ST >::gather_tables(), SplineC2ROMPTarget< ST >::gather_tables(), BsplineReader::initialize_spo2band(), MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalHamiltonianPool::make_hamWithEE(), MinimalWaveFunctionPool::make_O2_spinor(), MinimalWaveFunctionPool::make_O2_spinor_J12(), MinimalHamiltonianPool::makeHamWithEEEI(), makePsets(), MCPopulation::measureGlobalEnergyVariance(), mpiTestFunctionWrapped(), output_hardware_info(), TimerManager< qmcplusplus::TimerType< CLOCK > >::output_timing(), TimerManager< qmcplusplus::TimerType< CLOCK > >::print(), TimerManager< qmcplusplus::TimerType< CLOCK > >::print_flat(), TimerManager< qmcplusplus::TimerType< CLOCK > >::print_stack(), PWOrbitalSetBuilder::PWOrbitalSetBuilder(), QMCDriverNew::QMCDriverNew(), QMCWFOptLinearFactoryNew(), RandomNumberControl::read(), RandomNumberControl::read_parallel(), RandomNumberControl::read_rank_0(), hdf_archive::set_access_plist(), SetupSFNBranch::SetupSFNBranch(), SetupSimpleFixedNodeBranch::SetupSimpleFixedNodeBranch(), QMCDriverNew::setWalkerOffsets(), WalkerControl::syncFutureWalkersPerRank(), MCPopulation::syncWalkersPerRank(), TEST_CASE(), TraceManager::TraceManager(), WalkerLogManager::WalkerLogManager(), RandomNumberControl::write(), RandomNumberControl::write_parallel(), and RandomNumberControl::write_rank_0().
| std::atomic<size_t> CUDAallocator_device_mem_allocated |
|
static |
Definition at line 23 of file test_output_manager.cpp.
Referenced by init_string_output(), reset_string_output(), and TEST_CASE().
| std::vector<int> dims = {10, 10} |
Definition at line 51 of file test_ObservableHelper.cpp.
Referenced by Array< T, D >::Array(), checkShapeConsistency(), Array< T, D >::full_size(), h5d_append(), h5d_write(), EshdfFile::handleDensity(), EshdfFile::handleKpt(), HDFWalkerInput_0_4::read_hdf5(), HDFWalkerInput_0_4::read_hdf5_scatter(), HDFWalkerInput_0_4::read_phdf5(), Array< T, D >::resize(), IOVarASCII< T, RANK >::Resize(), ObservableHelper::set_dimensions(), TEST_CASE(), WalkerLogBuffer< WLog::Real >::writeHDF(), EshdfFile::writeQboxAtoms(), EshdfFile::writeQboxElectrons(), EshdfFile::writeQboxSupercell(), EshdfFile::writeQEAtoms(), and EshdfFile::writeQESupercell().
| const unsigned int DMAX = 4 |
Definition at line 47 of file TraceManager.h.
Referenced by TraceSample< TraceReal >::check_shape().
| TimerNameList_t<DMC_MPI_Timers> DMCMPITimerNames |
Definition at line 41 of file WalkerControlMPI.cpp.
| TimerNameList_t< DMCTimers > DMCTimerNames |
Definition at line 35 of file DMCUpdatePbyPFast.cpp.
|
static |
Definition at line 39 of file DensityMatrices1B.cpp.
| Libxml2Document doc |
Definition at line 368 of file test_OneBodyDensityMatrices.cpp.
Referenced by create_CN_Hamiltonian(), create_CN_particlesets(), qmcplusplus::testing::createEstimatorManagerNewGlobalInputXML(), qmcplusplus::testing::createEstimatorManagerNewInputXML(), qmcplusplus::testing::createEstimatorManagerNewVMCInputXML(), doSOECPotentialTest(), QMCGaussianParserBase::dump(), RMGParser::dumpPBC(), QMCGaussianParserBase::dumpPBC(), MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalParticlePool::make_H2(), MinimalHamiltonianPool::make_hamWithEE(), MinimalWaveFunctionPool::make_O2_spinor(), MinimalWaveFunctionPool::make_O2_spinor_J12(), MinimalHamiltonianPool::makeHamWithEEEI(), makeTestRPI(), output_hardware_info(), ParticleSetPool::output_particleset_info(), TimerManager< qmcplusplus::TimerType< CLOCK > >::output_timing(), parse_electron_ion_pbc_z(), parse_pbc_fcc_lattice(), parse_pbc_lattice(), MinimalParticlePool::parseParticleSetXML(), saveCusp(), HamiltonianPool::setDocument(), setup_He_wavefunction(), setupParticleSetPool(), setupParticleSetPoolBe(), SpaceGridEnv< VALID >::SpaceGridEnv(), SpaceGridEnv< ValidSpaceGridInput::valid::CYLINDRICAL >::SpaceGridEnv(), test_C_diamond(), test_cartesian_ao(), TEST_CASE(), TEST_CASE(), test_diamond_2x1x1_xml_input(), test_dirac_ao(), test_EtOH_mw(), test_HCN(), test_hcpBe_rotation(), test_He(), test_He_mw(), test_He_sto3g_xml_input(), test_J1_spline(), test_J3_polynomial3D(), test_LCAO_DiamondC_2x1x1_cplx(), test_LCAO_DiamondC_2x1x1_real(), test_lcao_spinor(), test_lcao_spinor_excited(), test_lcao_spinor_ion_derivs(), test_LiH_msd(), test_LiH_msd_xml_input(), test_LiH_msd_xml_input_with_positron(), test_Ne(), VMCBatchedTest::testCalcDefaultLocalWalkers(), and testTrialWaveFunction_diamondC_2x1x1().
| std::atomic<size_t> dual_device_mem_allocated |
| constexpr bool dump_obdm = false |
Definition at line 51 of file test_OneBodyDensityMatrices.cpp.
Referenced by TEST_CASE().
| testing::EstimatorManagerNewTest embt | ( | ham | , |
| c | , | ||
| 1 | |||
| ) |
Referenced by if(), and TEST_CASE().
Referenced by TEST_CASE().
| EstimatorManagerNewTestAccess emnta(emn) |
| EstimatorManagerNewTestAccess emnta2(emn2) |
|
static |
Definition at line 22 of file test_output_manager.cpp.
Referenced by init_string_output(), reset_string_output(), and TEST_CASE().
| Libxml2Document estimators_doc = createEstimatorManagerNewVMCInputXML() |
| Libxml2Document estimators_doc2 = createEstimatorManagerNewInputXML() |
Definition at line 77 of file test_EstimatorManagerNew.cpp.
| RNGThreadSafe<FakeRandom<OHMMS_PRECISION_FULL> > fake_random_global |
Definition at line 36 of file RandomGenerator.cpp.
| constexpr bool generate_test_data = false |
set to true to generate new random R for particles.
EXPECT all tests to fail in this case!
Definition at line 32 of file test_QMCHamiltonian.cpp.
Referenced by TEST_CASE(), OneBodyDensityMatricesTests< T >::testAccumulate(), and OneBodyDensityMatricesTests< T >::testEvaluateMatrix().
| std::vector<QMCTraits::RealType> good_data = embt.generateGoodOperatorData(num_ranks) |
Definition at line 97 of file test_manager_mpi.cpp.
| QMCHamiltonian ham = *(hamiltonian_pool.getPrimary()) |
Definition at line 38 of file test_EstimatorManagerNew.cpp.
Referenced by VMCBatched::advanceWalkers(), Crowd::Crowd(), EstimatorManagerNew::EstimatorManagerNew(), fill_from_text(), QMCDriverNew::initialLogEvaluation(), CloneManager::makeClones(), QMCHamiltonian::mw_evaluate(), QMCHamiltonian::mw_evaluateWithToperator(), SetupSFNBranch::operator()(), RMCLocalEnergyEstimator::resizeBasedOnHamiltonian(), RMCLocalEnergyEstimator::RMCLocalEnergyEstimator(), DMCBatched::run(), Crowd::setRNGForHamiltonian(), TEST_CASE(), QMCHamiltonian::updateComponent(), QMCHamiltonian::updateKinetic(), and QMCMain::validateXML().
| auto hamiltonian_pool = MinimalHamiltonianPool::make_hamWithEE(comm, particle_pool, wavefunction_pool) |
Definition at line 63 of file test_EstimatorManagerNew.cpp.
Referenced by QMCDriverFactory::createQMCDriver(), TEST_CASE(), and VMCBatchedTest::testCalcDefaultLocalWalkers().
| hdf_archive hFile |
Definition at line 45 of file test_ObservableHelper.cpp.
| hdf_error_suppression hide_hdf_errors |
Suppress HDF5 warning and error messages.
Definition at line 23 of file hdf_archive.cpp.
| auto & hstream = cuda_handles->hstream |
Definition at line 218 of file test_cuBLAS_LU.cpp.
Referenced by TEST_CASE().
| int iattribute = species_set.addAttribute("membersize") |
Definition at line 138 of file test_SpinDensityNew.cpp.
Referenced by qmcplusplus::testing::makeSpeciesSet(), and TEST_CASE().
| const std::string identity_coeff |
Definition at line 218 of file test_RotatedSPOs_LCAO.cpp.
Referenced by TEST_CASE().
| const int inone = std::numeric_limits<int>::min() |
Definition at line 23 of file SPOSetInputInfo.cpp.
Referenced by SPOSetInputInfo::put(), and SPOSetInputInfo::reset().
| int ispecies = species_set.addSpecies("C") |
Definition at line 137 of file test_SpinDensityNew.cpp.
Referenced by test_cartesian_ao(), TEST_CASE(), test_dirac_ao(), test_He(), test_He_mw(), test_He_sto3g_xml_input(), test_J1_spline(), test_LCAO_DiamondC_2x1x1_cplx(), test_LCAO_DiamondC_2x1x1_real(), test_lcao_spinor(), test_lcao_spinor_excited(), test_lcao_spinor_ion_derivs(), test_LiH_msd(), test_LiH_msd_xml_input(), test_LiH_msd_xml_input_with_positron(), and test_Ne().
| auto lattice = testing::makeTestLattice() |
Definition at line 142 of file test_SpinDensityNew.cpp.
Referenced by KContainer::BuildKLists(), SpinDensityInput::calculateDerivedParameters(), MagnetizationDensityInput::calculateDerivedParameters(), FreeOrbitalBuilder::createSPOSetFromXML(), doSOECPotentialTest(), SoaAtomicBasisSet< ROT, SH >::evaluateV(), SoaAtomicBasisSet< ROT, SH >::evaluateVGH(), SoaAtomicBasisSet< ROT, SH >::evaluateVGHGH(), SoaAtomicBasisSet< ROT, SH >::evaluateVGL(), KContainer::findApproxMMax(), makePsets(), qmcplusplus::testing::makeTestLattice(), SoaAtomicBasisSet< ROT, SH >::mw_evaluateV(), parse_pbc_fcc_lattice(), parse_pbc_lattice(), QMCDriverNew::QMCDriverNew(), test_C_diamond(), TEST_CASE(), test_CoulombPBCAA_3p(), test_diamond_2x1x1_xml_input(), test_hcpBe_rotation(), test_LCAO_DiamondC_2x1x1_cplx(), test_LCAO_DiamondC_2x1x1_real(), testTrialWaveFunction_diamondC_2x1x1(), and KContainer::updateKLists().
| int lda = 4 |
Definition at line 217 of file test_cuBLAS_LU.cpp.
Referenced by qmcplusplus::compute::applyW_batched(), qmcplusplus::SYCL::applyW_batched(), DiracMatrixComputeOMPTarget< VALUE_FP >::computeInvertAndLog(), DiracMatrix< VALUE_FP >::computeInvertAndLog(), computeLogDet_sycl(), qmcplusplus::SYCL::copyAinvRow_saveGL_batched(), qmcplusplus::compute::copyAinvRow_saveGL_batched(), qmcplusplus::cuBLAS::geam(), LAPACK::geev(), qmcplusplus::compute::BLAS::gemm(), BLAS::gemm(), qmcplusplus::syclBLAS::gemm(), qmcplusplus::ompBLAS::gemm< double >(), qmcplusplus::ompBLAS::gemm< float >(), qmcplusplus::ompBLAS::gemm< std::complex< double > >(), qmcplusplus::ompBLAS::gemm< std::complex< float > >(), qmcplusplus::compute::BLAS::gemm_batched(), qmcplusplus::ompBLAS::gemm_batched< double >(), qmcplusplus::ompBLAS::gemm_batched< float >(), qmcplusplus::ompBLAS::gemm_batched< std::complex< double > >(), qmcplusplus::ompBLAS::gemm_batched< std::complex< float > >(), qmcplusplus::ompBLAS::gemm_batched_impl(), qmcplusplus::ompBLAS::gemm_impl(), qmcplusplus::syclBLAS::gemv(), qmcplusplus::compute::BLAS::gemv(), BLAS::gemv(), qmcplusplus::ompBLAS::gemv< double >(), qmcplusplus::ompBLAS::gemv< float >(), qmcplusplus::ompBLAS::gemv< std::complex< double > >(), qmcplusplus::ompBLAS::gemv< std::complex< float > >(), qmcplusplus::compute::BLAS::gemv_batched(), qmcplusplus::syclBLAS::gemv_batched< double >(), qmcplusplus::ompBLAS::gemv_batched< double >(), qmcplusplus::syclBLAS::gemv_batched< float >(), qmcplusplus::ompBLAS::gemv_batched< float >(), qmcplusplus::syclBLAS::gemv_batched< std::complex< double > >(), qmcplusplus::ompBLAS::gemv_batched< std::complex< double > >(), qmcplusplus::syclBLAS::gemv_batched< std::complex< float > >(), qmcplusplus::ompBLAS::gemv_batched< std::complex< float > >(), qmcplusplus::ompBLAS::gemv_batched_impl(), qmcplusplus::ompBLAS::gemv_impl(), qmcplusplus::compute::BLAS::ger(), BLAS::ger(), qmcplusplus::ompBLAS::ger< double >(), qmcplusplus::ompBLAS::ger< float >(), qmcplusplus::ompBLAS::ger< std::complex< double > >(), qmcplusplus::ompBLAS::ger< std::complex< float > >(), qmcplusplus::compute::BLAS::ger_batched(), qmcplusplus::syclBLAS::ger_batched< double >(), qmcplusplus::ompBLAS::ger_batched< double >(), qmcplusplus::syclBLAS::ger_batched< float >(), qmcplusplus::ompBLAS::ger_batched< float >(), qmcplusplus::syclBLAS::ger_batched< std::complex< double > >(), qmcplusplus::ompBLAS::ger_batched< std::complex< double > >(), qmcplusplus::syclBLAS::ger_batched< std::complex< float > >(), qmcplusplus::ompBLAS::ger_batched< std::complex< float > >(), qmcplusplus::syclBLAS::ger_batched_impl(), qmcplusplus::ompBLAS::ger_batched_impl(), qmcplusplus::ompBLAS::ger_impl(), LAPACK::gesvd(), qmcplusplus::cusolver::getrf(), qmcplusplus::rocsolver::getrf(), qmcplusplus::cuBLAS::getrf_batched(), qmcplusplus::cusolver::getrf_bufferSize(), qmcplusplus::rocsolver::getri(), qmcplusplus::cuBLAS::getri_batched(), qmcplusplus::cusolver::getrs(), qmcplusplus::rocsolver::getrs(), LAPACK::ggev(), LAPACK::heev(), hipblasCgemmBatched(), hipblasCgetrfBatched_(), hipblasCgetriBatched_(), hipblasDgetrfBatched_(), hipblasDgetriBatched_(), hipblasSgetrfBatched_(), hipblasSgetriBatched_(), hipblasZgemmBatched(), hipblasZgetrfBatched_(), hipblasZgetriBatched_(), DiracMatrixComputeOMPTarget< VALUE_FP >::invert_transpose(), DiracMatrix< VALUE_FP >::invert_transpose(), DiracMatrixComputeCUDA< VALUE_FP >::invert_transpose(), DelayedUpdateBatched< PL, VALUE >::mw_accept_rejectRow(), DiracMatrixComputeCUDA< VALUE_FP >::mw_computeInvertAndLog(), DiracMatrixComputeCUDA< VALUE_FP >::mw_computeInvertAndLog_stride(), DiracMatrixComputeCUDA< VALUE_FP >::mw_invertTranspose(), DelayedUpdateBatched< PL, VALUE >::mw_updateInvMat(), DelayedUpdateBatched< PL, VALUE >::mw_updateRow(), PosSoA2AoS(), qmcplusplus::simd::remapCopy(), DiracMatrixComputeOMPTarget< VALUE_FP >::reset(), DiracMatrix< VALUE_FP >::reset(), TEST_CASE(), qmcplusplus::simd::transpose(), qmcplusplus::syclBLAS::transpose(), DelayedUpdateBatched< PL, VALUE >::updateRow(), Xgetrf(), and Xgetri().
| std::unordered_map<std::string, std::any> lookup_input_enum_value |
Definition at line 43 of file test_InputSection.cpp.
Referenced by TestInputSection::assignAnyEnum().
| std::vector< double > lu |
Definition at line 224 of file test_cuBLAS_LU.cpp.
Referenced by TEST_CASE().
| std::vector< double > lu2 |
Definition at line 241 of file test_cuBLAS_LU.cpp.
| lus[1] = dev_lu.data() |
Definition at line 264 of file test_cuBLAS_LU.cpp.
Referenced by TEST_CASE().
| std::vector<double, CUDAHostAllocator<double> > M2_vec {6, 5, 7, 5, 2, 2, 5, 4, 8, 2, 6, 4, 3, 8, 6, 8} |
Definition at line 380 of file test_cuBLAS_LU.cpp.
| std::vector<double, CUDAHostAllocator<double> > M_vec {2, 5, 7, 5, 5, 2, 5, 4, 8, 2, 6, 4, 7, 8, 6, 8} |
Definition at line 379 of file test_cuBLAS_LU.cpp.
Referenced by TEST_CASE().
| MCCoords<CoordsType::POS> mc_coords_rs(size_test) |
| MCCoords<CoordsType::POS_SPIN> mc_coords_rsspins(size_test) |
| std::vector<double*, CUDAHostAllocator<double*> > Ms {devM_vec.data(), devM2_vec.data()} |
Definition at line 383 of file test_cuBLAS_LU.cpp.
Referenced by TEST_CASE().
| int n = 4 |
Definition at line 216 of file test_cuBLAS_LU.cpp.
Referenced by qmcplusplus::simd::accumulate_n(), qmcplusplus::simd::accumulate_phases(), qmcplusplus::simd::add(), EstimatorManagerBase::add(), Matrix< ST, qmcplusplus::Mallocator< ST > >::add(), SpeciesSet::addAttribute(), XmlNode::addAttribute(), XmlNode::addParameterChild(), XmlNode::addYesNoAttribute(), XmlNode::addYesNoParameterChild(), Mallocator< RealType * >::allocate(), CUDAManagedAllocator< T >::allocate(), SYCLSharedAllocator< T, ALIGN >::allocate(), DualAllocator< T, DeviceAllocator, HostAllocator >::allocate(), OMPallocator< Value >::allocate(), CUDAAllocator< T_FP >::allocate(), SYCLAllocator< std::int64_t >::allocate(), CUDAHostAllocator< T_FP >::allocate(), SYCLHostAllocator< int >::allocate(), CUDALockedPageAllocator< T, ULPHA >::allocate(), IOVarBase::Append(), DTD_BConds< T, 3, PPPO >::apply_bc(), DTD_BConds< T, D, SC >::apply_bc(), DTD_BConds< T, 2, SUPERCELL_BULK >::apply_bc(), DTD_BConds< T, 3, PPPS >::apply_bc(), DTD_BConds< T, 2, PPPS >::apply_bc(), DTD_BConds< T, 2, PPPO >::apply_bc(), DTD_BConds< T, 3, PPPG >::apply_bc(), DTD_BConds< T, 2, SUPERCELL_WIRE >::apply_bc(), DTD_BConds< T, 3, PPNG >::apply_bc(), DTD_BConds< T, 3, PPNO >::apply_bc(), DTD_BConds< T, 3, PPNS >::apply_bc(), DTD_BConds< T, 3, SUPERCELL_WIRE >::apply_bc(), DTD_BConds< T, 3, PPPX >::apply_bc(), DTD_BConds< T, 3, PPNX >::apply_bc(), assignGaussRand(), assignUniformRand(), Matrix< ST, qmcplusplus::Mallocator< ST > >::attachReference(), Vector< T, std::allocator< T > >::attachReference(), VectorSoaContainer< ST, 5 >::attachReference(), BLAS::axpy(), MultiFunctorAdapter< FN >::batched_evaluate(), Communicate::bcast(), BranchIO< SFNB >::bcast_state(), EinsplineSetBuilder::bcastSortBands(), bspline(), TestSmallMatrixDetCalculator< T >::build_interal_data(), QMCFiniteSize::build_spherical_grid(), MultiDiracDeterminant::buildTableMatrix_calculateRatios_impl(), MultiDiracDeterminant::buildTableMatrix_calculateRatiosValueMatrixOneParticle(), LPQHIBasis::c(), LPQHISRCoulombBasis::c(), qmcplusplus::compute::calcGradients_batched(), qmcplusplus::SYCL::calcGradients_batched(), calcSmallDeterminant(), ci_configuration2::calculateExcitations(), ci_configuration2::calculateNumOfExcitations(), Cartesian2Spherical(), CartesianTensor< T, Point_t, Tensor_t, GGG_t >::CartesianTensor(), J1OrbitalSoA< FT >::checkOutVariables(), WalkerLogCollector::collect(), WalkerLogBuffer< WLog::Real >::collect(), DescentEngine::computeFinalizationUncertainties(), DiracMatrixComputeOMPTarget< VALUE_FP >::computeInvertAndLog(), DiracMatrix< VALUE_FP >::computeInvertAndLog(), computeLogDet(), computeLogDet_sycl(), conjdot(), DTD_BConds< T, 3, PPPX >::convert2Cart(), DTD_BConds< T, 3, PPPX >::convert2Unit(), convertToReal(), qmcplusplus::simd::copy(), BLAS::copy(), qmcplusplus::ompBLAS::copy< double >(), qmcplusplus::ompBLAS::copy< float >(), qmcplusplus::ompBLAS::copy< std::complex< double > >(), qmcplusplus::ompBLAS::copy< std::complex< float > >(), qmcplusplus::compute::BLAS::copy_batched(), qmcplusplus::ompBLAS::copy_batched< double >(), qmcplusplus::ompBLAS::copy_batched< float >(), qmcplusplus::ompBLAS::copy_batched< std::complex< double > >(), qmcplusplus::ompBLAS::copy_batched< std::complex< float > >(), qmcplusplus::ompBLAS::copy_batched_impl(), qmcplusplus::ompBLAS::copy_batched_offset< double >(), qmcplusplus::ompBLAS::copy_batched_offset< float >(), qmcplusplus::ompBLAS::copy_batched_offset< std::complex< double > >(), qmcplusplus::ompBLAS::copy_batched_offset< std::complex< float > >(), qmcplusplus::ompBLAS::copy_batched_offset_impl(), qmcplusplus::ompBLAS::copy_impl(), qmcplusplus::SYCL::copyAinvRow_saveGL_batched(), qmcplusplus::compute::copyAinvRow_saveGL_batched(), CUDAAllocator< T_FP >::copyDeviceToDevice(), SYCLAllocator< std::int64_t >::copyDeviceToDevice(), CUDAAllocator< T_FP >::copyFromDevice(), SYCLAllocator< std::int64_t >::copyFromDevice(), CUDAAllocator< T_FP >::copyToDevice(), SYCLAllocator< std::int64_t >::copyToDevice(), SPOSetInfo::count_degeneracies(), QMCGaussianParserBase::createCenter(), QMCGaussianParserBase::createCenterH5(), QMCGaussianParserBase::createDeterminantSet(), QMCGaussianParserBase::createDeterminantSetWithHDF5(), QMCGaussianParserBase::createMultiDeterminantSetCIHDF5(), QMCGaussianParserBase::createShell(), QMCGaussianParserBase::createShellH5(), QMCGaussianParserBase::createSPOSets(), QMCGaussianParserBase::createSPOSetsH5(), WalkerConfigurations::createWalkers(), MCWalkerConfiguration::createWalkers(), CubicSplineSolve(), CUDAfill_n(), TensorSoaContainer< T, 3 >::data(), LPQHISRCoulombBasis::dc_dk(), LPQHI_BasisClass::dDminus_dk(), LPQHI_BasisClass::dDplus_dk(), Mallocator< RealType * >::deallocate(), DualAllocator< T, DeviceAllocator, HostAllocator >::deallocate(), OMPallocator< Value >::deallocate(), CUDAAllocator< T_FP >::deallocate(), SYCLAllocator< std::int64_t >::deallocate(), CUDALockedPageAllocator< T, ULPHA >::deallocate(), LPQHI_BasisClass::dEminus_dk(), LPQHI_BasisClass::dEplus_dk(), Determinant(), qmc_allocator_traits< DualAllocator< T, DeviceAllocator, HostAllocator > >::deviceSideCopyN(), qmc_allocator_traits< OMPallocator< T, HostAllocator > >::deviceSideCopyN(), LRBasis::df_dr(), LRBasis::dfk_dk(), LPQHISRCoulombBasis::dh_ddelta(), LPQHIBasis::dh_dr(), LPQHISRCoulombBasis::dh_dr(), LPQHIBasis::Dminus(), LPQHISRCoulombBasis::Dminus(), LPQHI_BasisClass::Dminus(), LPQHISRCoulombBasis::Dminus_dG(), LRBreakup< BreakupBasis >::DoAllBreakup(), LRBreakup< BreakupBasis >::DoBreakup(), LRBreakup< BreakupBasis >::DoGradBreakup(), LRBreakup< BreakupBasis >::DoStrainBreakup(), qmcplusplus::simd::dot(), BLAS::dot(), LPQHIBasis::Dplus(), LPQHISRCoulombBasis::Dplus(), LPQHI_BasisClass::Dplus(), LPQHISRCoulombBasis::Dplus_dG(), LPQHIBasis::Eminus(), LPQHISRCoulombBasis::Eminus(), LPQHI_BasisClass::Eminus(), LPQHISRCoulombBasis::Eminus_dG(), LPQHIBasis::Eplus(), LPQHISRCoulombBasis::Eplus(), LPQHI_BasisClass::Eplus(), LPQHISRCoulombBasis::Eplus_dG(), error(), eval_e2iphi(), LRRPABFeeHandlerTemp< Func, BreakupBasis >::evalFk(), LRRPAHandlerTemp< Func, BreakupBasis >::evalFk(), LRHandlerTemp< Func, BreakupBasis >::evalFk(), MultiFunctorAdapter< FN >::evaluate(), L2Potential::evaluate(), LRRPABFeeHandlerTemp< Func, BreakupBasis >::evaluate(), LRHandlerTemp< Func, BreakupBasis >::evaluate(), LRRPAHandlerTemp< Func, BreakupBasis >::evaluate(), SmallMatrixDetCalculator< ValueType >::evaluate(), ShortRangeCuspFunctor< T >::evaluate(), CustomizedMatrixDet< 1 >::evaluate(), CustomizedMatrixDet< 2 >::evaluate(), PolynomialFunctor3D::evaluate(), SHOSet::evaluate_check(), DensityMatrices1B::evaluate_check(), SHOSet::evaluate_d0(), SHOSet::evaluate_d1(), SHOSet::evaluate_d2(), SHOSet::evaluate_hermite(), DensityMatrices1B::evaluate_loop(), DensityMatrices1B::evaluate_matrix(), CompositeSPOSet::evaluate_notranspose(), DTD_BConds< T, 3, PPPO >::evaluate_rsquared(), DTD_BConds< T, D, SC >::evaluate_rsquared(), DTD_BConds< T, 3, PPPS >::evaluate_rsquared(), DTD_BConds< T, 3, PPPG >::evaluate_rsquared(), DTD_BConds< T, 3, PPNG >::evaluate_rsquared(), DTD_BConds< T, 3, PPNO >::evaluate_rsquared(), DTD_BConds< T, 3, PPNS >::evaluate_rsquared(), DTD_BConds< T, 3, SUPERCELL_WIRE >::evaluate_rsquared(), DTD_BConds< T, 3, PPPX >::evaluate_rsquared(), DTD_BConds< T, 3, PPNX >::evaluate_rsquared(), MultiSlaterDetTableMethod::evaluate_vgl_impl(), DiracDeterminantWithBackflow::evaluateDerivatives(), ShortRangeCuspFunctor< T >::evaluateDerivatives(), BsplineFunctor< REAL >::evaluateDerivatives(), PolynomialFunctor3D::evaluateDerivatives(), TwoBodyJastrow< FT >::evaluateDerivativesWF(), TwoBodyJastrow< FT >::evaluateDerivRatios(), LRHandlerTemp< Func, BreakupBasis >::evaluateLR(), LRHandlerTemp< Func, BreakupBasis >::evaluateLR_r0(), OneBodyDensityMatrices::evaluateMatrix(), kSpaceJastrow::evaluateRatiosAlltoOne(), LRHandlerSRCoulomb< Func, BreakupBasis >::evaluateSR_k0(), LRHandlerTemp< Func, BreakupBasis >::evaluateSR_k0(), LRHandlerSRCoulomb< Func, BreakupBasis >::evaluateSR_k0_dstrain(), PolynomialFunctor3D::evaluateV(), CompositeSPOSet::evaluateValue(), CompositeSPOSet::evaluateVGL(), PolynomialFunctor3D::evaluateVGL(), CompositeSPOSet::evaluateVGL_spin(), RotatedSPOs::exponentiate_antisym_matrix(), LRBasis::f(), qmc_allocator_traits< Allocator >::fill_n(), qmc_allocator_traits< DualAllocator< T, DeviceAllocator, HostAllocator > >::fill_n(), qmc_allocator_traits< OMPallocator< T, HostAllocator > >::fill_n(), qmc_allocator_traits< qmcplusplus::CUDAAllocator< T > >::fill_n(), LRBasis::fk(), flat_idx(), PooledMemory< FullPrecRealType >::forward(), Communicate::gatherv(), qmcplusplus::cuBLAS::geam(), LAPACK::geev(), qmcplusplus::compute::BLAS::gemm(), qmcplusplus::syclBLAS::gemm(), qmcplusplus::compute::BLAS::gemm_batched(), qmcplusplus::ompBLAS::gemm_batched_impl(), qmcplusplus::ompBLAS::gemm_impl(), qmcplusplus::syclBLAS::gemv(), BLAS::gemv(), qmcplusplus::compute::BLAS::gemv(), qmcplusplus::ompBLAS::gemv< double >(), qmcplusplus::ompBLAS::gemv< float >(), qmcplusplus::ompBLAS::gemv< std::complex< double > >(), qmcplusplus::ompBLAS::gemv< std::complex< float > >(), qmcplusplus::compute::BLAS::gemv_batched(), qmcplusplus::syclBLAS::gemv_batched< double >(), qmcplusplus::ompBLAS::gemv_batched< double >(), qmcplusplus::syclBLAS::gemv_batched< float >(), qmcplusplus::ompBLAS::gemv_batched< float >(), qmcplusplus::syclBLAS::gemv_batched< std::complex< double > >(), qmcplusplus::ompBLAS::gemv_batched< std::complex< double > >(), qmcplusplus::syclBLAS::gemv_batched< std::complex< float > >(), qmcplusplus::ompBLAS::gemv_batched< std::complex< float > >(), qmcplusplus::ompBLAS::gemv_batched_impl(), qmcplusplus::ompBLAS::gemv_impl(), BLAS::gemv_trans(), DensityMatrices1B::generate_density_samples(), DensityMatrices1B::generate_particle_basis(), DensityMatrices1B::generate_sample_ratios(), OneBodyDensityMatrices::generateParticleBasis(), OneBodyDensityMatrices::generateSampleRatios(), qmcplusplus::compute::BLAS::ger(), BLAS::ger(), qmcplusplus::ompBLAS::ger< double >(), qmcplusplus::ompBLAS::ger< float >(), qmcplusplus::ompBLAS::ger< std::complex< double > >(), qmcplusplus::ompBLAS::ger< std::complex< float > >(), qmcplusplus::compute::BLAS::ger_batched(), qmcplusplus::syclBLAS::ger_batched< double >(), qmcplusplus::ompBLAS::ger_batched< double >(), qmcplusplus::syclBLAS::ger_batched< float >(), qmcplusplus::ompBLAS::ger_batched< float >(), qmcplusplus::syclBLAS::ger_batched< std::complex< double > >(), qmcplusplus::ompBLAS::ger_batched< std::complex< double > >(), qmcplusplus::syclBLAS::ger_batched< std::complex< float > >(), qmcplusplus::ompBLAS::ger_batched< std::complex< float > >(), qmcplusplus::syclBLAS::ger_batched_impl(), qmcplusplus::ompBLAS::ger_batched_impl(), qmcplusplus::ompBLAS::ger_impl(), LAPACK::gesvd(), PrimeNumberSet< uint_type >::get(), CartesianTensor< T, Point_t, Tensor_t, GGG_t >::getABC(), SoaCartesianTensor< T >::getABC(), getAlignedSize(), getBestTile(), GamesAsciiParser::getCSFSign(), GaussianFCHKParser::getGaussianCenters(), DiracParser::getGaussianCenters(), LCAOHDFParser::getMO(), qmcplusplus::cusolver::getrf(), qmcplusplus::rocsolver::getrf(), LAPACK::getrf(), qmcplusplus::cuBLAS::getrf_batched(), qmcplusplus::cusolver::getrf_bufferSize(), qmcplusplus::rocsolver::getri(), LAPACK::getri(), qmcplusplus::cuBLAS::getri_batched(), qmcplusplus::cusolver::getrs(), qmcplusplus::rocsolver::getrs(), getStats(), LAPACK::ggev(), GKIntegration< F, GKRule >::GK(), GKIntegration< F, GKRule >::GKGeneral(), LPQHIBasis::h(), LPQHISRCoulombBasis::h(), LAPACK::heev(), LPQHIBasis::hintr2(), LPQHISRCoulombBasis::hintr2(), hipblasCgemmBatched(), hipblasCgetrfBatched_(), hipblasCgetriBatched_(), hipblasDgetrfBatched_(), hipblasDgetriBatched_(), hipblasSgetrfBatched_(), hipblasSgetriBatched_(), hipblasZgemmBatched(), hipblasZgetrfBatched_(), hipblasZgetriBatched_(), Crowd::incNonlocalAccept(), BsplineSet::init_base(), SHOSet::initialize(), SpaceGrid::initialize_rectilinear(), RMCUpdateAllWithDrift::initWalkers(), RMCUpdatePbyPWithDrift::initWalkers(), DensityMatrices1B::integrate(), qmcplusplus::simd::inv(), Invert(), invert_matrix(), DiracMatrixComputeOMPTarget< VALUE_FP >::invert_transpose(), DiracMatrix< VALUE_FP >::invert_transpose(), DiracMatrixComputeCUDA< VALUE_FP >::invert_transpose(), InvertLU(), InvertWithLog(), PooledMemory< FullPrecRealType >::lendReference(), Limits(), LimitsInclusive(), RotatedSPOs::log_antisym_matrix(), LRHandlerTemp< Func, BreakupBasis >::lrDf(), LUFactorization(), RandomNumberControl::make_children(), make_seed(), J1OrbitalSoA< FT >::makeClone(), qmcplusplus::testing::makeRngSpdMatrix(), qmcplusplus::testing::makeRngVector(), Matrix< ST, qmcplusplus::Mallocator< ST > >::Matrix(), MCSample::MCSample(), DescentEngine::mpi_unbiased_ratio_of_means(), DiracMatrixComputeCUDA< VALUE_FP >::mw_computeInvertAndLog(), DiracMatrixComputeCUDA< VALUE_FP >::mw_computeInvertAndLog_stride(), DiracMatrixComputeCUDA< VALUE_FP >::mw_invertTranspose(), norm(), BLAS::norm2(), Normalize(), SPOSetInputInfo::occupy_energies(), OneDimQuinticSpline< Td, Tg, CTd, CTg >::OneDimQuinticSpline(), TinyVector< T, N, base_type >::operator()(), STONorm< T >::operator()(), TestFillBufferRngReal< T >::operator()(), TestFillVecRngReal< T >::operator()(), TestGetVecRngReal< T >::operator()(), TestFillBufferRngComplex< T >::operator()(), TestFillVecRngComplex< T >::operator()(), TestGetVecRngComplex< T >::operator()(), OrthogExcluding(), OrthogLower(), Orthogonalize(), Orthogonalize2(), OutOfPlaceTranspose(), parseGridInput(), PooledData< RealType >::PooledData(), pow(), PrimeNumberSet< uint_type >::PrimeNumberSet(), print(), QMCFiniteSize::printSkRawSphAvg(), qmcplusplus::MatrixOperators::product(), OrbitalImages::put(), putContent(), LCAOSpinorBuilder::putFromH5(), LCAOrbitalBuilder::putFromH5(), LCAOrbitalBuilder::putFromXML(), LCAOrbitalBuilder::putPBCFromH5(), RecordNamedProperty< RealType >::RecordNamedProperty(), Communicate::reduce(), Communicate::reduce_in_place(), SpaceGrid::registerCollectables(), NESpaceGrid< REAL >::registerGrid(), qmcplusplus::simd::remainder(), qmcplusplus::simd::remapCopy(), OrbitalImages::report(), BlockHistogram< T >::reserve(), PooledData< RealType >::reserve(), DiracMatrixComputeOMPTarget< VALUE_FP >::reset(), GaussianTimesRN< T >::reset(), PolynomialFunctor3D::reset_gamma(), container_traits< Vector< T, ALLOC > >::resize(), container_traits< boost::multi::array< T, D, Alloc > >::resize(), RealSpacePositions::resize(), SmallMatrixDetCalculator< ValueType >::resize(), BlockHistogram< T >::resize(), RealSpacePositionsOMPTarget::resize(), container_traits< std::vector< T, ALLOC > >::resize(), container_traits< Matrix< T, ALLOC > >::resize(), TensorSoaContainer< T, 3 >::resize(), PolynomialFunctor3D::resize(), container_traits< Array< T, D > >::resize(), PooledData< RealType >::resize(), OneDimGridFunctor< T, T, Vector< T >, Vector< T > >::resize(), PairCorrEstimator::resize(), Matrix< ST, qmcplusplus::Mallocator< ST > >::resize(), BsplineFunctor< REAL >::resize(), IOVarASCII< T, RANK >::Resize(), container_proxy< std::vector< T > >::resize(), VectorSoaContainer< ST, 5 >::resize(), ConstantSizeMatrix< FullPrecRealType, std::allocator< FullPrecRealType > >::resize(), Vector< T, std::allocator< T > >::resize(), container_proxy< PooledData< T > >::resize(), RecordNamedProperty< RealType >::resize(), container_proxy< Vector< T > >::resize(), container_proxy< Matrix< T > >::resize(), NumericalGrid< T, CT >::resize(), SymmArray< T >::resize(), Vector< T, std::allocator< T > >::resize_impl(), SOECPComponent::resize_warrays(), NonLocalECPComponent::resize_warrays(), Walker< qmcplusplus::QMCTraits, qmcplusplus::PtclOnLatticeTraits >::resizeProperty(), SplineC2R< ST >::resizeStorage(), SplineR2R< ST >::resizeStorage(), SplineC2C< ST >::resizeStorage(), SplineC2COMPTarget< ST >::resizeStorage(), SplineC2ROMPTarget< ST >::resizeStorage(), SimpleGrid::ReverseMap(), LPQHISRCoulombBasis::rh(), DensityMatrices1B::same(), BLAS::scal(), STONorm< T >::set(), LogGridLight< T >::set(), OneDimQuinticSpline< Td, Tg, CTd, CTg >::set(), LinearGrid< RealType >::set(), LogGrid< T, CT >::set(), LogGridZero< T, CT >::set(), NumericalGrid< T, CT >::set(), PairCorrEstimator::set_norm_factor(), LPQHIBasis::set_NumKnots(), LPQHISRCoulombBasis::set_NumKnots(), CubicBsplineGrid< T, LINEAR_1DGRID, FIRSTDERIV_CONSTRAINTS >::setGrid(), SampleStack::setMaxSamples(), Communicate::setNumNodes(), OneDimGridFunctor< T, T, Vector< T >, Vector< T > >::setNumOfNodes(), MCWalkerConfiguration::setNumSamples(), QMCGaussianParserBase::setOccupationNumbers(), BackflowFunctionBase::setParamIndex(), RecordPropertyList::setstride(), SpeciesSet::setTotalNum(), OhmmsAsciiParser::skiplines(), SoaCartesianTensor< T >::SoaCartesianTensor(), qmcplusplus::simd::sqrt(), LRRPABFeeHandlerTemp< Func, BreakupBasis >::srDf(), LRRPAHandlerTemp< Func, BreakupBasis >::srDf(), LRHandlerTemp< Func, BreakupBasis >::srDf(), SpaceGrid::sum(), NESpaceGrid< REAL >::sum(), DescentEngine::takeSample(), TensorSoaContainer< T, 3 >::TensorSoaContainer(), RandomNumberControl::test(), TEST_CASE(), SHOSet::test_derivatives(), test_J1_spline(), SHOSet::test_overlap(), testDualAllocator(), qmcplusplus::simd::transpose(), Transpose(), qmcplusplus::syclBLAS::transpose(), EinsplineSetBuilder::TwistPair(), SHOSetBuilder::update_basis_states(), qmc_allocator_traits< DualAllocator< T, DeviceAllocator, HostAllocator > >::updateFrom(), qmc_allocator_traits< qmcplusplus::SYCLAllocator< T > >::updateFrom(), qmc_allocator_traits< DualAllocator< T, DeviceAllocator, HostAllocator > >::updateTo(), qmc_allocator_traits< qmcplusplus::SYCLAllocator< T > >::updateTo(), TraceSamples< std::complex< TraceReal > >::user_report(), Vector< T, std::allocator< T > >::Vector(), VectorSoaContainer< ST, 5 >::VectorSoaContainer(), EnergyDensityEstimator::write_nonzero_domains(), TraceRequest::write_selected(), WalkerLogManager::writeBuffers(), WalkerLogBuffer< WLog::Real >::writeSummary(), Xgetrf(), and Xgetri().
Definition at line 23 of file SPOSetInfo.cpp.
Referenced by SPOSetInfo::modify().
| constexpr int no_index = -1 |
Definition at line 24 of file SPOSetInfo.cpp.
Referenced by SPOSetInfo::modify().
| xmlNodePtr node = doc.getRoot() |
Definition at line 372 of file test_OneBodyDensityMatrices.cpp.
Referenced by Libxml2Document::addChild(), EstimatorManagerInput::appendEstimatorInput(), EstimatorManagerInput::appendScalarEstimatorInput(), qmcplusplus::testing::createDriver(), qmcplusplus::testing::createEstimatorManagerNewGlobalInputXML(), qmcplusplus::testing::createEstimatorManagerNewInputXML(), qmcplusplus::testing::createEstimatorManagerNewVMCInputXML(), makeTestRPI(), DMCBatched::process(), VMCDriverInput::readXML(), DMCDriverInput::readXML(), SpaceGridEnv< VALID >::SpaceGridEnv(), SpaceGridEnv< ValidSpaceGridInput::valid::CYLINDRICAL >::SpaceGridEnv(), SpinDensityInput::SpinDensityInput(), TrialWaveFunction::storeXMLNode(), TEST_CASE(), EstimatorManagerInputTests::testAppendFromXML(), and VMCBatchedTest::testCalcDefaultLocalWalkers().
| int num_ranks = c->size() |
Definition at line 68 of file test_manager_mpi.cpp.
Referenced by QMCDriverNew::adjustGlobalWalkerCount(), determineDefaultDeviceNum(), EstimatorManagerBaseTest::EstimatorManagerBaseTest(), EstimatorManagerNewTest::EstimatorManagerNewTest(), FakeOperatorEstimator::FakeOperatorEstimator(), EstimatorManagerNewTest::generateGoodOperatorData(), if(), SampleStack::setMaxSamples(), TEST_CASE(), and UnifiedDriverWalkerControlMPITest::UnifiedDriverWalkerControlMPITest().
| OneBodyDensityMatricesInput obdmi(node) |
Referenced by EstimatorManagerNew::put(), and TEST_CASE().
| oeba[0] = 1.0 |
Definition at line 162 of file test_SpinDensityNew.cpp.
Referenced by TEST_CASE().
| int offset_for_rs = (3 * size_test) + (3 * size_test) % 2 |
Definition at line 138 of file test_random_seq.cpp.
| ObservableHelper oh {hdf_path{"u"}} |
Definition at line 50 of file test_ObservableHelper.cpp.
Referenced by EnergyDensityEstimator::registerCollectables(), StaticStructureFactor::registerCollectables(), SpaceGrid::registerCollectables(), SpinDensity::registerCollectables(), DensityMatrices1B::registerCollectables(), NESpaceGrid< REAL >::registerGrid(), OperatorBase::registerObservables(), MagnetizationDensity::registerOperatorEstimator(), SpinDensityNew::registerOperatorEstimator(), OneBodyDensityMatrices::registerOperatorEstimator(), ReferencePoints::save(), and TEST_CASE().
| bool okay = doc.parseFromString(Input::xml[Input::valid::VANILLA]) |
Definition at line 369 of file test_OneBodyDensityMatrices.cpp.
Referenced by create_CN_Hamiltonian(), create_CN_particlesets(), qmcplusplus::testing::createEstimatorManagerNewGlobalInputXML(), qmcplusplus::testing::createEstimatorManagerNewInputXML(), qmcplusplus::testing::createEstimatorManagerNewVMCInputXML(), doSOECPotentialTest(), MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalWaveFunctionPool::make_O2_spinor(), MinimalWaveFunctionPool::make_O2_spinor_J12(), makeTestRPI(), parse_electron_ion_pbc_z(), parse_pbc_fcc_lattice(), parse_pbc_lattice(), h5data_proxy< bool >::read(), ECPComponentBuilder::read_pp_file(), setup_He_wavefunction(), setupParticleSetPool(), setupParticleSetPoolBe(), SpaceGridEnv< VALID >::SpaceGridEnv(), SpaceGridEnv< ValidSpaceGridInput::valid::CYLINDRICAL >::SpaceGridEnv(), test_C_diamond(), test_cartesian_ao(), TEST_CASE(), TEST_CASE(), test_diamond_2x1x1_xml_input(), test_dirac_ao(), test_EtOH_mw(), test_HCN(), test_hcpBe_rotation(), test_He(), test_He_mw(), test_He_sto3g_xml_input(), test_J1_spline(), test_J3_polynomial3D(), test_LCAO_DiamondC_2x1x1_cplx(), test_LCAO_DiamondC_2x1x1_real(), test_lcao_spinor(), test_lcao_spinor_excited(), test_lcao_spinor_ion_derivs(), test_LiH_msd(), test_LiH_msd_xml_input(), test_LiH_msd_xml_input_with_positron(), test_Ne(), OneBodyDensityMatricesTests< T >::testRegisterAndWrite(), and testTrialWaveFunction_diamondC_2x1x1().
| std::atomic<size_t> OMPallocator_device_mem_allocated |
| const std::map<std::string, OptimizerType> OptimizerNames |
Definition at line 30 of file OptimizerTypes.h.
Referenced by QMCFixedSampleLinearOptimize::processOptXML(), QMCFixedSampleLinearOptimizeBatched::processOptXML(), and HybridEngine::processXML().
| SpinDensityNew original(std::move(sdi), lattice, species_set) |
| auto particle_pool = MinimalParticlePool::make_diamondC_1x1x1(comm) |
Definition at line 59 of file test_EstimatorManagerNew.cpp.
Referenced by QMCDriverFactory::createQMCDriver(), MinimalWaveFunctionPool::make_diamondC_1x1x1(), MinimalHamiltonianPool::make_hamWithEE(), MinimalWaveFunctionPool::make_O2_spinor(), MinimalWaveFunctionPool::make_O2_spinor_J12(), MinimalHamiltonianPool::makeHamWithEEEI(), makePsets(), TEST_CASE(), and VMCBatchedTest::testCalcDefaultLocalWalkers().
| std::vector<int, CUDAHostAllocator<int> > pivots = {3, 4, 3, 4, 3, 4, 4, 4} |
Definition at line 271 of file test_cuBLAS_LU.cpp.
Referenced by TEST_CASE().
| SetupPools pools |
Definition at line 82 of file test_SFNBranch.cpp.
Referenced by CrowdWithWalkers::CrowdWithWalkers(), and TEST_CASE().
| float propertyFloat = 10.f |
Definition at line 52 of file test_ObservableHelper.cpp.
| Matrix<float> propertyMatrix |
Definition at line 58 of file test_ObservableHelper.cpp.
Definition at line 55 of file test_ObservableHelper.cpp.
| TinyVector<float, OHMMS_DIM> propertyTinyVector |
Definition at line 61 of file test_ObservableHelper.cpp.
| std::vector<float> propertyVector |
Definition at line 64 of file test_ObservableHelper.cpp.
| std::vector<TinyVector<float, OHMMS_DIM> > propertyVectorTinyVector |
Definition at line 67 of file test_ObservableHelper.cpp.
| auto& pset = *(particle_pool.getParticleSet("e")) |
Definition at line 62 of file test_EstimatorManagerNew.cpp.
Referenced by MagnetizationDensity::accumulate(), SelfHealingOverlap::accumulate(), SpinDensityNew::accumulate(), MomentumDistribution::accumulate(), accumulateFromPsets(), TestSOECPotential::addVPs(), VMCBatched::advanceWalkers(), QMCUpdateBase::checkLogAndGL(), WalkerLogCollector::collect(), Crowd::Crowd(), EstimatorManagerNew::EstimatorManagerNew(), PSdispatcher::flex_donePbyP(), PSdispatcher::flex_update(), ParticleSetPool::get(), QMCDriverNew::initialLogEvaluation(), SampleStack::loadSample(), MCWalkerConfiguration::loadSample(), makePsets(), ParticleSet::mw_accept_rejectMove(), ParticleSet::mw_donePbyP(), SoaDistanceTableABOMPTarget< T, D, SC >::mw_evaluate(), TrialWaveFunction::mw_evaluateDeltaLog(), TrialWaveFunction::mw_evaluateDeltaLogSetup(), TrialWaveFunction::mw_evaluateGL(), TrialWaveFunction::mw_evaluateLog(), BareKineticEnergy::mw_evaluatePerParticle(), CoulombPBCAA::mw_evaluatePerParticle(), CoulombPBCAB::mw_evaluatePerParticle(), ParticleSet::mw_loadWalker(), ParticleSet::mw_update(), NEReferencePoints::NEReferencePoints(), NonLocalECPComponent::NonLocalECPComponent(), SetupSFNBranch::operator()(), NEReferencePoints::processParticleSets(), EstimatorManagerNew::put(), randomUpdateAccumulate(), ParticleSetPool::reset(), TEST_CASE(), OneBodyDensityMatricesTests< T >::testEvaluateMatrix(), QMCHamiltonian::updateComponent(), and QMCHamiltonian::updateKinetic().
| auto& pset_target = *(particle_pool.getParticleSet("e")) |
Definition at line 364 of file test_OneBodyDensityMatrices.cpp.
Referenced by OneBodyDensityMatrices::calcDensity(), OneBodyDensityMatrices::calcDensityDrift(), OneBodyDensityMatrices::calcDensityDriftWithSpin(), OneBodyDensityMatrices::calcDensityWithSpin(), OneBodyDensityMatrices::evaluateMatrix(), OneBodyDensityMatrices::generateDensitySamples(), OneBodyDensityMatrices::generateDensitySamplesWithSpin(), OneBodyDensityMatrices::generateParticleBasis(), qmcplusplus::testing::generateRandomParticleSets(), OneBodyDensityMatrices::generateSampleBasis(), OneBodyDensityMatrices::generateSampleRatios(), OneBodyDensityMatrices::generateSamples(), MagnetizationDensity::generateSpinIntegrand(), OneBodyDensityMatrices::normalizeBasis(), OneBodyDensityMatrices::OneBodyDensityMatrices(), EstimatorManagerNew::put(), TEST_CASE(), OneBodyDensityMatricesTests< T >::testGenerateSamples(), OneBodyDensityMatricesTests< T >::testGenerateSamplesForSpinor(), OneBodyDensityMatrices::updateBasis(), OneBodyDensityMatrices::updateBasisD012(), OneBodyDensityMatrices::updateBasisD012WithSpin(), OneBodyDensityMatrices::updateBasisWithSpin(), and OneBodyDensityMatrices::warmupSampling().
| QMCState qmc_common |
a unique QMCState during a run
Definition at line 111 of file qmc_common.cpp.
Referenced by kSpaceJastrowBuilder::buildComponent(), QMCMain::execute(), QMCMain::executeLoop(), main(), CloneManager::makeClones(), kSpaceJastrowBuilder::outputJastrow(), ProjectData::previousRoot(), ECPComponentBuilder::printECPTable(), VMC::put(), CSVMC::put(), QMCDriver::putQMCInfo(), QMCMain::QMCMain(), EinsplineSetBuilder::ReadOrbitalInfo_ESHDF(), ProjectData::reset(), VMC::resetRun(), and QMCMain::validateXML().
| RNGThreadSafe< RandomGenerator > random_global |
Definition at line 37 of file RandomGenerator.cpp.
| std::vector<int> real_pivot {3, 3, 4, 4, 3, 3, 3, 4} |
Definition at line 439 of file test_cuBLAS_LU.cpp.
Referenced by TEST_CASE().
Definition at line 24 of file SPOSetInputInfo.cpp.
Referenced by SPOSetInputInfo::find_energy_extrema(), SPOSetInputInfo::put(), and SPOSetInputInfo::reset().
| RunTimeManager< ChronoClock > run_time_manager |
Definition at line 25 of file RunTimeManager.cpp.
Referenced by QMCMain::execute(), QMCMain::executeLoop(), VMC::run(), RMC::run(), DMC::run(), VMCBatched::run(), and DMCBatched::run().
| SpinDensityInput sdi | ( | node | ) |
Referenced by TEST_CASE().
| SpinDensityInput sdi_copy = sdi |
Definition at line 140 of file test_SpinDensityNew.cpp.
| SpinDensityNew sdn(std::move(sdi), lattice, species_set) |
Referenced by accumulateFromPsets(), TEST_CASE(), and SpinDensityNewTests::testCopyConstructor().
| SpinDensityNewTests sdnt |
Definition at line 163 of file test_SpinDensityNew.cpp.
| SetupSimpleFixedNodeBranch setup_sfnb |
Definition at line 112 of file test_SimpleFixedNodeBranch.cpp.
Definition at line 86 of file test_SFNBranch.cpp.
Referenced by SetupSFNBranch::createMyNode(), SetupSimpleFixedNodeBranch::createMyNode(), SetupSFNBranch::operator()(), and SetupSimpleFixedNodeBranch::operator()().
| const std::string& snone = "none" |
Definition at line 25 of file SPOSetInputInfo.cpp.
Referenced by SPOSetInputInfo::put(), and SPOSetInputInfo::reset().
| const TimerNameList_t< SODMCTimers > SODMCTimerNames |
Definition at line 31 of file SODMCUpdatePbyPFast.cpp.
| species_set = pset_target.getSpeciesSet() |
Definition at line 365 of file test_OneBodyDensityMatrices.cpp.
Referenced by qmcplusplus::testing::makeSpeciesSet(), and TEST_CASE().
| auto& spomap = wavefunction_pool.getWaveFunction("wavefunction")->getSPOMap() |
Definition at line 366 of file test_OneBodyDensityMatrices.cpp.
Referenced by OneBodyDensityMatrices::OneBodyDensityMatrices(), DensityMatrices1B::set_state(), TrialWaveFunction::storeSPOMap(), and TEST_CASE().
|
static |
Definition at line 44 of file TrialWaveFunction.cpp.
Referenced by TrialWaveFunction::addComponent(), create_names(), and TrialWaveFunction::TrialWaveFunction().
|
static |
Definition at line 20 of file test_output_manager.cpp.
Referenced by init_string_output(), reset_string_output(), and TEST_CASE().
| std::atomic<size_t> SYCLallocator_device_mem_allocated |
| ProjectData test_project("test", ProjectData::DriverVersion::BATCH) |
Referenced by SetupPools::SetupPools(), TEST_CASE(), and test_hcpBe_rotation().
| TimerNameList_t<TestTimer> TestTimerNames = {{MyTimer1, "Timer name 1"}, {MyTimer2, "Timer name 2"}} |
Definition at line 397 of file test_timer.cpp.
Referenced by TEST_CASE().
| bool timer_max_level_exceeded = false |
Definition at line 26 of file NewTimer.cpp.
Referenced by StackKeyParam< 2 >::add_id(), TimerManager< qmcplusplus::TimerType< CLOCK > >::output_timing(), and TimerManager< qmcplusplus::TimerType< CLOCK > >::print_stack().
| auto& twf = *(wavefunction_pool.getWaveFunction("wavefunction")) |
Definition at line 64 of file test_EstimatorManagerNew.cpp.
Referenced by WalkerConsumer::addWalker(), Crowd::addWalker(), VMCBatched::advanceWalkers(), DMCBatched::advanceWalkers(), QMCUpdateBase::checkLogAndGL(), Crowd::Crowd(), EstimatorManagerNew::EstimatorManagerNew(), TrialWaveFunction::initializeTWFFastDerivWrapper(), QMCDriverNew::initialLogEvaluation(), TrialWaveFunction::mw_evaluateDeltaLog(), TrialWaveFunction::mw_evaluateDeltaLogSetup(), TrialWaveFunction::mw_evaluateGL(), TrialWaveFunction::mw_evaluateLog(), SetupSFNBranch::operator()(), EstimatorManagerNew::put(), SlaterDet::registerTWFFastDerivWrapper(), DiracDeterminant< DU_TYPE >::registerTWFFastDerivWrapper(), DiracDeterminantBatched< PL, VT, FPVT >::registerTWFFastDerivWrapper(), TEST_CASE(), and test_LiH_msd().
| TimerNameList_t<WC_Timers> WalkerControlTimerNames |
Definition at line 47 of file WalkerControl.cpp.
| auto wavefunction_pool |
Definition at line 60 of file test_EstimatorManagerNew.cpp.
Referenced by QMCDriverFactory::createQMCDriver(), MinimalHamiltonianPool::make_hamWithEE(), MinimalHamiltonianPool::makeHamWithEEEI(), TEST_CASE(), and VMCBatchedTest::testCalcDefaultLocalWalkers().